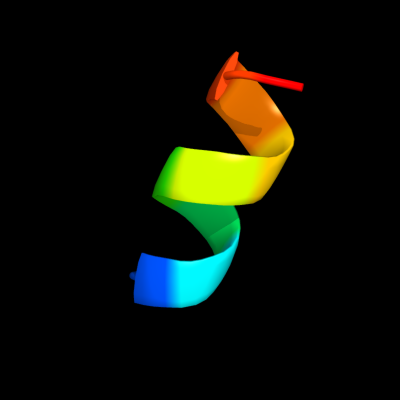| 1 |
|
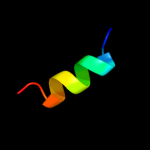
PDB 1r7m chain A domain 1
Region: 76 - 85
Aligned: 10
Modelled: 10
Confidence: 15.7%
Identity: 40%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 2 |
|
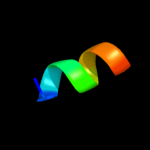
PDB 2lkg chain A
Region: 116 - 153
Aligned: 38
Modelled: 38
Confidence: 10.2%
Identity: 11%
PDB header:signaling protein
Chain: A: PDB Molecule:acetylcholine receptor;
PDBTitle: wsa major conformation
Phyre2
| 3 |
|
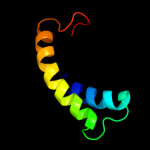
PDB 3cm1 chain C
Region: 146 - 161
Aligned: 16
Modelled: 16
Confidence: 9.2%
Identity: 31%
PDB header:cell cycle
Chain: C: PDB Molecule:ssga-like sporulation-specific cell division protein;
PDBTitle: crystal structure of ssga-like sporulation-specific cell division2 protein (yp_290167.1) from thermobifida fusca yx-er1 at 2.60 a3 resolution
Phyre2
| 4 |
|
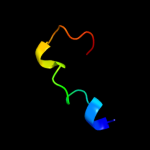
PDB 1fn9 chain A
Region: 143 - 159
Aligned: 17
Modelled: 17
Confidence: 7.6%
Identity: 12%
Fold: Outer capsid protein sigma 3
Superfamily: Outer capsid protein sigma 3
Family: Outer capsid protein sigma 3
Phyre2
| 5 |
|
PDB 2b3g chain B
Region: 1 - 14
Aligned: 14
Modelled: 14
Confidence: 7.5%
Identity: 14%
PDB header:replication
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53n (fragment 33-60) bound to rpa70n
Phyre2
| 6 |
|
PDB 2yvx chain D
Region: 73 - 140
Aligned: 67
Modelled: 68
Confidence: 6.6%
Identity: 24%
PDB header:transport protein
Chain: D: PDB Molecule:mg2+ transporter mgte;
PDBTitle: crystal structure of magnesium transporter mgte
Phyre2
| 7 |
|
PDB 1qlo chain A
Region: 93 - 106
Aligned: 14
Modelled: 14
Confidence: 6.3%
Identity: 14%
PDB header:membrane proteins
Chain: A: PDB Molecule:herpes simplex virus protein icp47;
PDBTitle: structure of the active domain of the herpes simplex virus2 protein icp47 in water/sodium dodecyl sulfate solution3 determined by nuclear magnetic resonance spectroscopy
Phyre2
| 8 |
|
PDB 1r7m chain A
Region: 76 - 85
Aligned: 10
Modelled: 10
Confidence: 6.2%
Identity: 40%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:intron-encoded endonuclease i-scei;
PDBTitle: the homing endonuclease i-scei bound to its dna recognition2 region
Phyre2
| 9 |
|
PDB 2kr6 chain A
Region: 159 - 213
Aligned: 55
Modelled: 55
Confidence: 6.1%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:presenilin-1;
PDBTitle: solution structure of presenilin-1 ctf subunit
Phyre2
| 10 |
|
PDB 2zfd chain B
Region: 100 - 121
Aligned: 22
Modelled: 22
Confidence: 5.7%
Identity: 18%
PDB header:signaling protein/transferase
Chain: B: PDB Molecule:putative uncharacterized protein t20l15_90;
PDBTitle: the crystal structure of plant specific calcium binding protein atcbl22 in complex with the regulatory domain of atcipk14
Phyre2
| 11 |
|
PDB 2d10 chain F
Region: 205 - 212
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 38%
PDB header:cell adhesion
Chain: F: PDB Molecule:ezrin-radixin-moesin binding phosphoprotein 50;
PDBTitle: crystal structure of the radixin ferm domain complexed with2 the nherf-1 c-terminal tail peptide
Phyre2
| 12 |
|
PDB 2d10 chain G
Region: 205 - 212
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 38%
PDB header:cell adhesion
Chain: G: PDB Molecule:ezrin-radixin-moesin binding phosphoprotein 50;
PDBTitle: crystal structure of the radixin ferm domain complexed with2 the nherf-1 c-terminal tail peptide
Phyre2
| 13 |
|
PDB 2d10 chain E
Region: 205 - 212
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 38%
PDB header:cell adhesion
Chain: E: PDB Molecule:ezrin-radixin-moesin binding phosphoprotein 50;
PDBTitle: crystal structure of the radixin ferm domain complexed with2 the nherf-1 c-terminal tail peptide
Phyre2
| 14 |
|
PDB 2d10 chain H
Region: 205 - 212
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 38%
PDB header:cell adhesion
Chain: H: PDB Molecule:ezrin-radixin-moesin binding phosphoprotein 50;
PDBTitle: crystal structure of the radixin ferm domain complexed with2 the nherf-1 c-terminal tail peptide
Phyre2