| 1 |
|
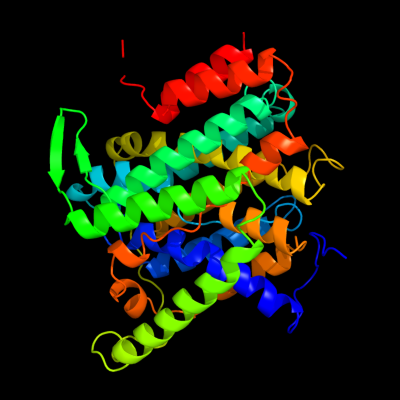
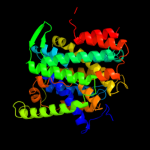
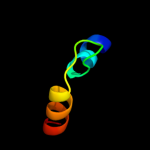
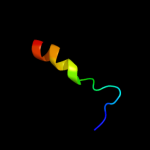
PDB 3qe7 chain A
Region: 18 - 418
Aligned: 385
Modelled: 399
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:uracil permease;
PDBTitle: crystal structure of uracil transporter--uraa
Phyre2
| 2 |
|
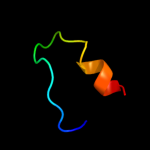
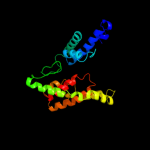
PDB 3lpz chain A
Region: 300 - 313
Aligned: 14
Modelled: 14
Confidence: 15.7%
Identity: 14%
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
Phyre2
| 3 |
|
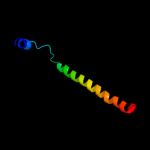
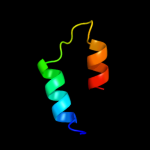
PDB 2dme chain A
Region: 2 - 36
Aligned: 32
Modelled: 35
Confidence: 15.5%
Identity: 16%
PDB header:metal binding protein
Chain: A: PDB Molecule:phd finger protein 3;
PDBTitle: solution structure of the tfiis domain ii of human phd2 finger protein 3
Phyre2
| 4 |
|
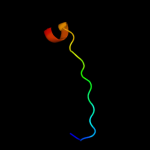
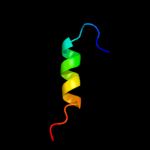
PDB 1w7c chain A domain 3
Region: 12 - 38
Aligned: 22
Modelled: 27
Confidence: 11.2%
Identity: 23%
Fold: Cystatin-like
Superfamily: Amine oxidase N-terminal region
Family: Amine oxidase N-terminal region
Phyre2
| 5 |
|
PDB 2elp chain A
Region: 15 - 35
Aligned: 19
Modelled: 21
Confidence: 10.9%
Identity: 16%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
Phyre2
| 6 |
|
PDB 3din chain D
Region: 372 - 422
Aligned: 51
Modelled: 51
Confidence: 10.1%
Identity: 10%
PDB header:membrane protein, protein transport
Chain: D: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 7 |
|
PDB 2jqo chain A
Region: 1 - 19
Aligned: 19
Modelled: 19
Confidence: 9.5%
Identity: 16%
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical protein yoba;
PDBTitle: nmr solution structure of bacillus subtilis yoba 21-120:2 northeast structural genomics consortium target sr547
Phyre2
| 8 |
|
PDB 1enw chain A
Region: 2 - 36
Aligned: 32
Modelled: 35
Confidence: 9.4%
Identity: 6%
Fold: RuvA C-terminal domain-like
Superfamily: Elongation factor TFIIS domain 2
Family: Elongation factor TFIIS domain 2
Phyre2
| 9 |
|
PDB 1w3g chain A
Region: 13 - 31
Aligned: 19
Modelled: 19
Confidence: 9.0%
Identity: 16%
PDB header:toxin/lectin
Chain: A: PDB Molecule:hemolytic lectin from laetiporus sulphureus;
PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
Phyre2
| 10 |
|
PDB 2axt chain F domain 1
Region: 417 - 441
Aligned: 25
Modelled: 25
Confidence: 7.9%
Identity: 24%
Fold: Single transmembrane helix
Superfamily: Cytochrome b559 subunits
Family: Cytochrome b559 subunits
Phyre2
| 11 |
|
PDB 3ndq chain A
Region: 2 - 36
Aligned: 32
Modelled: 32
Confidence: 7.8%
Identity: 22%
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor a protein 1;
PDBTitle: structure of human tfiis domain ii
Phyre2
| 12 |
|
PDB 1xsz chain A domain 2
Region: 1 - 19
Aligned: 19
Modelled: 19
Confidence: 6.9%
Identity: 16%
Fold: TBP-like
Superfamily: RalF, C-terminal domain
Family: RalF, C-terminal domain
Phyre2
| 13 |
|
PDB 3arc chain L
Region: 428 - 441
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 29%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 14 |
|
PDB 3e5a chain B
Region: 7 - 25
Aligned: 19
Modelled: 19
Confidence: 6.5%
Identity: 11%
PDB header:transferase
Chain: B: PDB Molecule:targeting protein for xklp2;
PDBTitle: crystal structure of aurora a in complex with vx-680 and tpx2
Phyre2
| 15 |
|
PDB 1p7g chain A domain 2
Region: 7 - 25
Aligned: 19
Modelled: 19
Confidence: 6.3%
Identity: 5%
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Superfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Phyre2
| 16 |
|
PDB 3er7 chain A domain 1
Region: 16 - 33
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 28%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: Exig0174-like
Phyre2
| 17 |
|
PDB 2ht2 chain B
Region: 14 - 223
Aligned: 199
Modelled: 210
Confidence: 5.9%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 18 |
|
PDB 3bu8 chain B
Region: 6 - 35
Aligned: 30
Modelled: 30
Confidence: 5.5%
Identity: 13%
PDB header:dna binding protein
Chain: B: PDB Molecule:telomeric repeat-binding factor 2;
PDBTitle: crystal structure of trf2 trfh domain and tin2 peptide2 complex
Phyre2
| 19 |
|
PDB 1dt0 chain A domain 2
Region: 7 - 26
Aligned: 20
Modelled: 20
Confidence: 5.4%
Identity: 15%
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Superfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), C-terminal domain
Phyre2
| 20 |
|
PDB 2aex chain A
Region: 3 - 26
Aligned: 24
Modelled: 24
Confidence: 5.4%
Identity: 13%
PDB header:oxidoreductase
Chain: A: PDB Molecule:coproporphyrinogen iii oxidase, mitochondrial;
PDBTitle: the 1.58a crystal structure of human coproporphyrinogen oxidase2 reveals the structural basis of hereditary coproporphyria
Phyre2
| 21 |
|