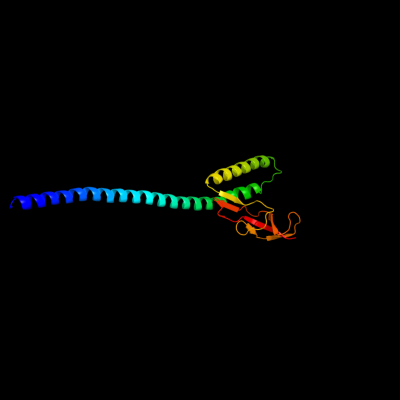
1 c1dkgB_
100.0
99
PDB header: complex (hsp24/hsp70)Chain: B: PDB Molecule: nucleotide exchange factor grpe;PDBTitle: crystal structure of the nucleotide exchange factor grpe2 bound to the atpase domain of the molecular chaperone dnak
2 c3a6mB_
100.0
27
PDB header: chaperoneChain: B: PDB Molecule: protein grpe;PDBTitle: crystal structure of grpe from thermus thermophilus hb8
3 d1dkga1
99.8
100
Fold: Head domain of nucleotide exchange factor GrpESuperfamily: Head domain of nucleotide exchange factor GrpEFamily: Head domain of nucleotide exchange factor GrpE4 c3hl6B_
27.8
22
PDB header: unknown functionChain: B: PDB Molecule: pathogenicity island protein;PDBTitle: staphylococcus aureus pathogenicity island 3 orf9 protein
5 c3swfA_
21.8
15
PDB header: transport proteinChain: A: PDB Molecule: cgmp-gated cation channel alpha-1;PDBTitle: cnga1 621-690 containing clz domain
6 d1rsoa_
19.6
14
Fold: L27 domainSuperfamily: L27 domainFamily: L27 domain7 d1nzea_
18.8
18
Fold: Four-helical up-and-down bundleSuperfamily: Oxygen-evolving enhancer protein 3,Family: Oxygen-evolving enhancer protein 3,8 c1nzeA_
18.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: oxygen-evolving enhancer protein 3;PDBTitle: crystal structure of psbq polypeptide of photosystem ii2 from higher plants
9 d1x5ta1
16.4
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD10 c2jvoA_
15.0
22
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: segmental isotope labeling of npl3
11 c1w5fA_
14.9
5
PDB header: cell divisionChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
12 d2cpia1
14.4
33
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD13 c3e7lD_
14.2
15
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
14 c3mkxC_
14.1
21
PDB header: antiviral proteinChain: C: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: crystal structure of bst2/tetherin
15 d1fnxh1
14.0
14
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD16 d2cbia1
13.5
16
Fold: Hyaluronidase domain-likeSuperfamily: Hyaluronidase post-catalytic domain-likeFamily: Hyaluronidase post-catalytic domain-like17 d1rk8a_
13.4
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD18 d1wi8a_
13.1
17
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD19 d1etxa_
12.6
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like20 d2choa1
12.6
10
Fold: Hyaluronidase domain-likeSuperfamily: Hyaluronidase post-catalytic domain-likeFamily: Hyaluronidase post-catalytic domain-like21 d2qeec1
not modelled
11.6
19
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Uronate isomerase-like22 d1w5fa2
not modelled
11.3
5
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain23 d1ntca_
not modelled
11.0
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like24 d1ttea1
not modelled
10.7
22
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain25 c3bs9A_
not modelled
10.6
17
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolysin tia-1 isoform p40;PDBTitle: x-ray structure of human tia-1 rrm2
26 d2j01s1
not modelled
9.8
30
Fold: Ribonuclease H-like motifSuperfamily: Translational machinery componentsFamily: Ribosomal protein L18 and S1127 c3lraA_
not modelled
9.5
16
PDB header: membrane proteinChain: A: PDB Molecule: disks large homolog 1, maguk p55 subfamily member 7,PDBTitle: structural basis for assembling a human tripartite complex dlg1-mpp7-2 mals3
28 d2cqca1
not modelled
9.5
21
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD29 d1we1a_
not modelled
9.1
6
Fold: Heme oxygenase-likeSuperfamily: Heme oxygenase-likeFamily: Eukaryotic type heme oxygenase30 c3he5A_
not modelled
9.0
24
PDB header: de novo proteinChain: A: PDB Molecule: synzip1;PDBTitle: heterospecific coiled-coil pair synzip2:synzip1
31 c2x7aB_
not modelled
8.8
21
PDB header: immune systemChain: B: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: structural basis of hiv-1 tethering to membranes by the2 bst2-tetherin ectodomain
32 d1fxla1
not modelled
8.6
13
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD33 d1pzqa_
not modelled
8.6
25
Fold: Dimerisation interlockSuperfamily: Docking domain A of the erythromycin polyketide synthase (DEBS)Family: Docking domain A of the erythromycin polyketide synthase (DEBS)34 c1zvuA_
not modelled
8.5
23
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase iv subunit a;PDBTitle: structure of the full-length e. coli parc subunit
35 c3movB_
not modelled
7.8
17
PDB header: structural proteinChain: B: PDB Molecule: lamin-b1;PDBTitle: crystal structure of human lamin-b1 coil 2 segment
36 c2jl8S_
not modelled
7.8
30
PDB header: ribosomeChain: S: PDB Molecule: 50s ribosomal protein l18;PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome (part 4 of 4).3 this file contains the 50s subunit.
37 d2d6fa1
not modelled
7.6
10
Fold: Sm-like foldSuperfamily: GatD N-terminal domain-likeFamily: GatD N-terminal domain-like38 c3q9vB_
not modelled
7.5
29
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding response regulator;PDBTitle: crystal structure of rra c-terminal domain(123-221) from deinococcus2 radiodurans
39 c3m9bK_
not modelled
7.4
11
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
40 d1h2vz_
not modelled
7.3
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD41 c3md1B_
not modelled
7.1
13
PDB header: rna binding proteinChain: B: PDB Molecule: nuclear and cytoplasmic polyadenylated rna-binding proteinPDBTitle: crystal structure of the second rrm domain of yeast poly(u)-binding2 protein (pub1)
42 d1igub_
not modelled
7.0
10
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Transcriptional repressor protein KorB43 d1fipa_
not modelled
6.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like44 d1tafa_
not modelled
6.7
7
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs45 d2zjrl1
not modelled
6.6
30
Fold: Ribonuclease H-like motifSuperfamily: Translational machinery componentsFamily: Ribosomal protein L18 and S1146 d1cvja1
not modelled
6.5
11
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD47 d2otaa1
not modelled
6.5
33
Fold: YejL-likeSuperfamily: YejL-likeFamily: YejL-like48 c3swyB_
not modelled
6.3
12
PDB header: transport proteinChain: B: PDB Molecule: cyclic nucleotide-gated cation channel alpha-3;PDBTitle: cnga3 626-672 containing clz domain
49 d1ovya_
not modelled
6.2
40
Fold: Ribonuclease H-like motifSuperfamily: Translational machinery componentsFamily: Ribosomal protein L18 and S1150 c2kxyA_
not modelled
6.1
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of sur18c from streptococcus thermophilus.2 northeast structural genomics consortium target sur18c
51 d2cqba1
not modelled
6.0
22
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD52 c1qzuB_
not modelled
5.9
25
PDB header: lyaseChain: B: PDB Molecule: hypothetical protein mds018;PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
53 d1igqa_
not modelled
5.9
17
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Transcriptional repressor protein KorB54 d1ov2a_
not modelled
5.9
13
Fold: RAP domain-likeSuperfamily: RAP domain-likeFamily: RAP domain55 c1xe1A_
not modelled
5.8
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pf0907;PDBTitle: hypothetical protein from pyrococcus furiosus pfu-880080-001
56 d1xe1a_
not modelled
5.8
31
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors57 d1bv8a_
not modelled
5.8
11
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Alpha-macroglobulin receptor domainFamily: Alpha-macroglobulin receptor domain58 d2j0sd1
not modelled
5.6
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD59 c1m7lA_
not modelled
5.5
11
PDB header: sugar binding proteinChain: A: PDB Molecule: pulmonary surfactant-associated protein d;PDBTitle: solution structure of the coiled-coil trimerization domain2 from lung surfactant protein d
60 d1kyza2
not modelled
5.4
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Plant O-methyltransferase, C-terminal domain61 d1jeyb1
not modelled
5.3
8
Fold: SPOC domain-likeSuperfamily: SPOC domain-likeFamily: Ku80 subunit middle domain62 d1huwa_
not modelled
5.2
12
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines63 c3cxjB_
not modelled
5.2
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from2 methanothermobacter thermautotrophicus
64 c3nmdA_
not modelled
5.2
17
PDB header: transferaseChain: A: PDB Molecule: cgmp dependent protein kinase;PDBTitle: crystal structure of the leucine zipper domain of cgmp dependent2 protein kinase i beta
65 d1jeya1
not modelled
5.2
17
Fold: SPOC domain-likeSuperfamily: SPOC domain-likeFamily: Ku70 subunit middle domain