| 1 |
|
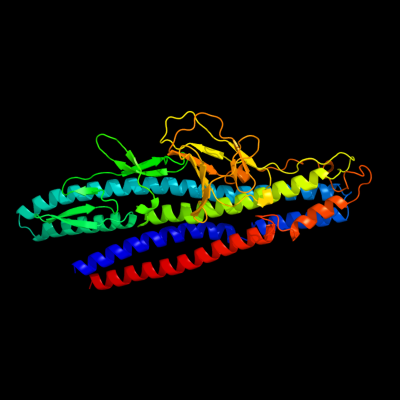
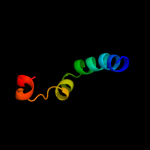
PDB 2d4y chain A
Region: 68 - 506
Aligned: 428
Modelled: 437
Confidence: 100.0%
Identity: 75%
PDB header:structural protein
Chain: A: PDB Molecule:flagellar hook-associated protein 1;
PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
Phyre2
| 2 |
|
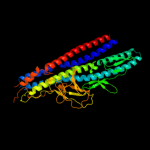
PDB 3a69 chain A
Region: 4 - 547
Aligned: 343
Modelled: 358
Confidence: 100.0%
Identity: 19%
PDB header:motor protein
Chain: A: PDB Molecule:flagellar hook protein flge;
PDBTitle: atomic model of the bacterial flagellar hook based on2 docking an x-ray derived structure and terminal two alpha-3 helices into an 7.1 angstrom resolution cryoem map
Phyre2
| 3 |
|
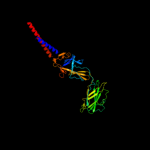
PDB 1wlg chain A
Region: 75 - 508
Aligned: 288
Modelled: 304
Confidence: 99.4%
Identity: 15%
Fold: Flagellar hook protein flgE
Superfamily: Flagellar hook protein flgE
Family: Flagellar hook protein flgE
Phyre2
| 4 |
|
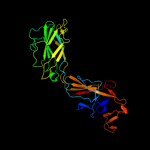
PDB 1ucu chain A
Region: 121 - 546
Aligned: 405
Modelled: 409
Confidence: 95.9%
Identity: 14%
Fold: Phase 1 flagellin
Superfamily: Phase 1 flagellin
Family: Phase 1 flagellin
Phyre2
| 5 |
|
PDB 3k8v chain B
Region: 470 - 529
Aligned: 60
Modelled: 60
Confidence: 88.6%
Identity: 18%
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
Phyre2
| 6 |
|
PDB 1ory chain B
Region: 510 - 546
Aligned: 37
Modelled: 37
Confidence: 55.5%
Identity: 19%
PDB header:chaperone
Chain: B: PDB Molecule:flagellin;
PDBTitle: flagellar export chaperone in complex with its cognate binding partner
Phyre2
| 7 |
|
PDB 2cos chain A
Region: 538 - 545
Aligned: 8
Modelled: 8
Confidence: 13.4%
Identity: 25%
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine protein kinase lats2;
PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
Phyre2
| 8 |
|
PDB 1y02 chain A
Region: 514 - 524
Aligned: 11
Modelled: 11
Confidence: 8.0%
Identity: 45%
PDB header:metal binding protein
Chain: A: PDB Molecule:fyve-ring finger protein sakura;
PDBTitle: crystal structure of a fyve-type domain from caspase2 regulator carp2
Phyre2
| 9 |
|
PDB 2c5r chain A domain 1
Region: 514 - 526
Aligned: 13
Modelled: 13
Confidence: 7.8%
Identity: 31%
Fold: Phage replication organizer domain
Superfamily: Phage replication organizer domain
Family: Phage replication organizer domain
Phyre2
| 10 |
|
PDB 1lvo chain A
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 7.1%
Identity: 13%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 11 |
|
PDB 2kxw chain B
Region: 514 - 526
Aligned: 13
Modelled: 13
Confidence: 7.0%
Identity: 23%
PDB header:calcium-binding protein/metal transport
Chain: B: PDB Molecule:sodium channel protein type 2 subunit alpha;
PDBTitle: structure of the c-domain fragment of apo calmodulin bound to the iq2 motif of nav1.2
Phyre2
| 12 |
|
PDB 2q6f chain B
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 6.9%
Identity: 7%
PDB header:hydrolase
Chain: B: PDB Molecule:infectious bronchitis virus (ibv) main protease;
PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
Phyre2
| 13 |
|
PDB 3kf6 chain B
Region: 29 - 51
Aligned: 23
Modelled: 23
Confidence: 6.8%
Identity: 26%
PDB header:structural protein
Chain: B: PDB Molecule:protein ten1;
PDBTitle: crystal structure of s. pombe stn1-ten1 complex
Phyre2
| 14 |
|
PDB 2duc chain A domain 1
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 6.5%
Identity: 13%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 15 |
|
PDB 1nrj chain A
Region: 26 - 34
Aligned: 9
Modelled: 9
Confidence: 6.4%
Identity: 22%
Fold: Profilin-like
Superfamily: SNARE-like
Family: SRP alpha N-terminal domain-like
Phyre2
| 16 |
|
PDB 2kgb chain I
Region: 536 - 547
Aligned: 12
Modelled: 12
Confidence: 5.5%
Identity: 42%
PDB header:contractile protein/ca binding protein
Chain: I: PDB Molecule:troponin i, cardiac muscle;
PDBTitle: nmr solution of the regulatory domain cardiac f77w-troponin2 c in complex with the cardiac troponin i 144-163 switch3 peptide
Phyre2
| 17 |
|
PDB 2l1r chain B
Region: 536 - 547
Aligned: 12
Modelled: 12
Confidence: 5.5%
Identity: 42%
PDB header:metal binding protein
Chain: B: PDB Molecule:troponin i;
PDBTitle: the structure of the calcium-sensitizer, dfbp-o, in complex with the2 n-domain of troponin c and the switch region of troponin i
Phyre2