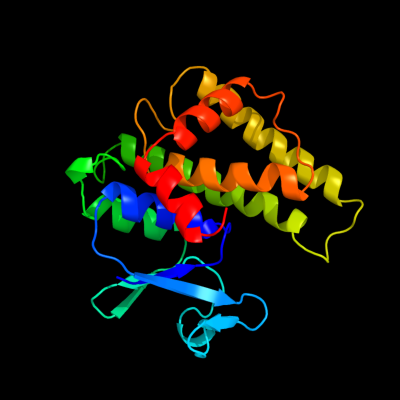
| 1 | c1f2eB_
|
|
 |
100.0 |
42 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: structure of sphingomonad, glutathione s-transferase complexed with2 glutathione
|
| 2 | c2pmtA_
|
|
 |
100.0 |
54 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione transferase;
PDBTitle: glutathione transferase from proteus mirabilis
|
| 3 | c2gdrA_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of a bacterial glutathione transferase
|
| 4 | c3uarA_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of glutathione transferase (target efi-501774) from2 methylococcus capsulatus str. bath with gsh bound
|
| 5 | c2ntoA_
|
|
 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: structure of the glutathione transferase from ochrobactrum anthropi in2 complex with glutathione
|
| 6 | c3c8eB_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:yghu, glutathione s-transferase homologue;
PDBTitle: crystal structure analysis of yghu from e. coli
|
| 7 | c1n2aB_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of a bacterial glutathione transferase2 from escherichia coli with glutathione sulfonate in the3 active site
|
| 8 | c2x64A_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione-s-transferase;
PDBTitle: glutathione-s-transferase from xylella fastidiosa
|
| 9 | c1ljrB_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: glutathione transferase (hgst t2-2) from human
|
| 10 | c1c72A_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:protein (glutathione s-transferase);
PDBTitle: tyr115, gln165 and trp209 contribute to the 1,2-epoxy-3-(p-2 nitrophenoxy)propane conjugating activities of glutathione3 s-transferase cgstm1-1
|
| 11 | c3m0fA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein gst_n;
PDBTitle: crystal structure of glutathione s transferase in complex2 with glutathione from pseudomonas fluorescens
|
| 12 | c3gx0A_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:gst-like protein yfcg;
PDBTitle: crystal structure of gsh-dependent disulfide bond2 oxidoreductase
|
| 13 | c2fheA_
|
|
 |
100.0 |
16 |
PDB header:transferase/substrate
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: fasciola hepatica glutathione s-transferase isoform 1 in complex with2 glutathione
|
| 14 | c1gtuB_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: ligand-free human glutathione s-transferase m1a-1a
|
| 15 | c1b8xA_
|
|
 |
100.0 |
14 |
PDB header:signal protein
Chain: A: PDB Molecule:protein (aml-1b);
PDBTitle: glutathione s-transferase fused with the nuclear matrix targeting2 signal of the transcription factor aml-1
|
| 16 | c3ergA_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase 2;
PDBTitle: crystal structure of gtt2 from saccharomyces cerevisiae in complex2 with glutathione sulfnate
|
| 17 | c3lykA_
|
|
 |
100.0 |
23 |
PDB header:transport protein
Chain: A: PDB Molecule:stringent starvation protein a homolog;
PDBTitle: structure of stringent starvation protein a homolog from2 haemophilus influenzae
|
| 18 | c3csiA_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase p;
PDBTitle: crystal structure of the glutathione transferase pi allelic variant*c,2 i104v/a113v, in complex with the chlorambucil-glutathione conjugate
|
| 19 | c3lszA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of glutathione s-transferase from2 rhodobacter sphaeroides
|
| 20 | c1ua5A_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: non-fusion gst from s. japonicum in complex with glutathione
|
| 21 | c1zl9A_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase 5;
PDBTitle: crystal structure of a major nematode c.elegans specific gst (ce01613)
|
| 22 | c1k3yB_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase a1;
PDBTitle: crystal structure analysis of human glutathione s-transferase with s-2 hexyl glutatione and glycerol at 1.3 angstrom
|
| 23 | c3qagA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase omega-2;
PDBTitle: human glutathione transferase o2 with glutathione -new crystal form
|
| 24 | c3isoB_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:putative glutathione transferase;
PDBTitle: crystal structure of 26 kda gst of clonorchis sinensis in p32212 symmetry
|
| 25 | c1tu8A_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase 2;
PDBTitle: structure of onchoverca volvulus pi-class glutathione s-2 transferase with its kompetitive inhibitor s-hexyl-gsh
|
| 26 | c1jlvA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione transferase gst1-3;
PDBTitle: anopheles dirus species b glutathione s-transferases 1-3
|
| 27 | c3m3mA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of glutathione s-transferase from pseudomonas2 fluorescens [pf-5]
|
| 28 | c1yq1A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: structural genomics of caenorhabditis elegans: glutathione2 s-transferase
|
| 29 | c2jl4A_ |
|
not modelled |
100.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:maleylpyruvate isomerase;
PDBTitle: holo structure of maleyl pyruvate isomerase, a bacterial2 glutathione-s-transferase in zeta class
|
| 30 | c1iyiA_ |
|
not modelled |
100.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:hematopoietic prostagladin d synthase;
PDBTitle: crystal structure of hematopoietic prostaglandin d synthase
|
| 31 | c2gsqA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: glutathione s-transferase from squid digestive gland complexed with s-2 (3-iodobenzyl)glutathione
|
| 32 | c3h1nA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable glutathione s-transferase;
PDBTitle: crystal structure of probable glutathione s-transferase from2 bordetella bronchiseptica rb50
|
| 33 | c1nhyA_ |
|
not modelled |
100.0 |
16 |
PDB header:translation
Chain: A: PDB Molecule:elongation factor 1-gamma 1;
PDBTitle: crystal structure of the gst-like domain of elongation2 factor 1-gamma from saccharomyces cerevisiae.
|
| 34 | c3f6fA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:cg18548-pa (ip02196p) (ip02193p);
PDBTitle: crystal structure of glutathione transferase dmgstd10 from2 drosophila melanogaster
|
| 35 | c1m0uB_ |
|
not modelled |
100.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:gst2 gene product;
PDBTitle: crystal structure of the drosophila glutathione s-2 transferase-2 in complex with glutathione
|
| 36 | c1yy7A_ |
|
not modelled |
100.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:stringent starvation protein a;
PDBTitle: crystal structure of stringent starvation protein a (sspa),2 an rna polymerase-associated transcription factor
|
| 37 | c1gumA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:protein (glutathione transferase a4-4);
PDBTitle: human glutathione transferase a4-4 without ligands
|
| 38 | c1vf1A_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase 3;
PDBTitle: cgsta1-1 in complex with glutathione
|
| 39 | c3lypA_ |
|
not modelled |
100.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:stringent starvation protein a;
PDBTitle: structure of stringent starvation protein a homolog from pseudomonas2 fluorescens
|
| 40 | c3nivD_ |
|
not modelled |
100.0 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:glutathione s-transferase;
PDBTitle: the crystal structure of glutathione s-transferase from legionella2 pneumophila
|
| 41 | c1oktA_ |
|
not modelled |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: x-ray structure of glutathione s-transferase from the2 malarial parasite plasmodium falciparum
|
| 42 | c1gwcC_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:glutathione s-transferase tsi-1;
PDBTitle: the structure of a tau class glutathione s-transferase from2 wheat, active in herbicide detoxification
|
| 43 | c1v2aD_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:glutathione transferase gst1-6;
PDBTitle: glutathione s-transferase 1-6 from anopheles dirus species b
|
| 44 | c3lxzD_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:glutathione s-transferase family protein;
PDBTitle: structure of probable glutathione s-transferase(pp0183) from2 pseudomonas putida
|
| 45 | c2vo4A_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:2,4-d inducible glutathione s-transferase;
PDBTitle: glutathione transferase from glycine max
|
| 46 | c2aheA_ |
|
not modelled |
100.0 |
19 |
PDB header:metal transport
Chain: A: PDB Molecule:chloride intracellular channel protein 4;
PDBTitle: crystal structure of a soluble form of clic4. intercellular2 chloride ion channel
|
| 47 | c1r5aA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione transferase;
PDBTitle: glutathione s-transferase
|
| 48 | c1oyjC_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure solution of rice gst1 (osgstu1) in complex with2 glutathione.
|
| 49 | c2hnlB_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase 1;
PDBTitle: structure of the prostaglandin d synthase from the parasitic nematode2 onchocerca volvulus
|
| 50 | c2fnoB_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:agr_pat_752p;
PDBTitle: crystal structure of a glutathione s-transferase (atu5508) from2 agrobacterium tumefaciens str. c58 at 2.00 a resolution
|
| 51 | c2c3nB_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase theta 1;
PDBTitle: human glutathione-s-transferase t1-1, apo form
|
| 52 | c2on7A_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:na glutathione s-transferase 1;
PDBTitle: structure of nagst-1
|
| 53 | c1gnwA_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: structure of glutathione s-transferase
|
| 54 | c1eemA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione-s-transferase;
PDBTitle: glutathione transferase from homo sapiens
|
| 55 | c1byeA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:protein (glutathione s-transferase);
PDBTitle: glutathione s-transferase i from mais in complex with2 atrazine glutathione conjugate
|
| 56 | c1jlwA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione transferase gst1-4;
PDBTitle: anopheles dirus species b glutathione s-transferases 1-4
|
| 57 | c2on5E_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: E: PDB Molecule:na glutathione s-transferase 2;
PDBTitle: structure of nagst-2
|
| 58 | c1bg5A_ |
|
not modelled |
100.0 |
14 |
PDB header:ankyrin binding
Chain: A: PDB Molecule:fusion protein of alpha-na,k-atpase with
PDBTitle: crystal structure of the ankyrin binding domain of alpha-na,2 k-atpase as a fusion protein with glutathione s-transferase
|
| 59 | c1k0dB_ |
|
not modelled |
100.0 |
22 |
PDB header:gene regulation
Chain: B: PDB Molecule:ure2 protein;
PDBTitle: ure2p in complex with glutathione
|
| 60 | c1g7oA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin 2;
PDBTitle: nmr solution structure of reduced e. coli glutaredoxin 2
|
| 61 | c2cz2A_ |
|
not modelled |
100.0 |
19 |
PDB header:isomerase, transferase
Chain: A: PDB Molecule:maleylacetoacetate isomerase;
PDBTitle: crystal structure of glutathione transferase zeta 1-12 (maleylacetoacetate isomerase) from mus musculus (form-1 crystal)
|
| 62 | c1b48A_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:protein (glutathione s-transferase);
PDBTitle: crystal structure of mgsta4-4 in complex with gsh conjugate2 of 4-hydroxynonenal in one subunit and gsh in the other:3 evidence of signaling across dimer interface in mgsta4-4
|
| 63 | c3lg6B_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:putative glutathione transferase;
PDBTitle: crystal structure of putative glutathione transferase from2 coccidioides immitis
|
| 64 | c3lq7B_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of glutathione s-transferase from2 agrobacterium tumefaciens str. c58
|
| 65 | c1aw9A_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase iii;
PDBTitle: structure of glutathione s-transferase iii in apo form
|
| 66 | c3bbyA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized gst-like protein yfcf;
PDBTitle: crystal structure of glutathione s-transferase (np_416804.1) from2 escherichia coli k12 at 1.85 a resolution
|
| 67 | c2wb9A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione transferase sigma class;
PDBTitle: fasciola hepatica sigma class gst
|
| 68 | c3ay8A_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: glutathione s-transferase unclassified 2 from bombyx mori
|
| 69 | c2imiA_ |
|
not modelled |
100.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:epsilon-class glutathione s-transferase;
PDBTitle: structures of an insect epsilon-class glutathione s-2 transferase from the malaria vector anopheles gambiae:3 evidence for high ddt-detoxifying activity
|
| 70 | c3m8nA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:possible glutathione s-transferase;
PDBTitle: crystal structure of a possible gutathione s-tranferase from2 rhodopseudomonas palustris
|
| 71 | c1k0nB_ |
|
not modelled |
100.0 |
18 |
PDB header:metal transport
Chain: B: PDB Molecule:chloride intracellular channel protein 1;
PDBTitle: chloride intracellular channel 1 (clic1) complexed with glutathione
|
| 72 | c2ws2B_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase;
PDBTitle: the 2 angstrom structure of a nu-class gst from haemonchus contortus
|
| 73 | c3cbuB_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:probable gst-related protein;
PDBTitle: crystal structure of a putative glutathione s-transferase (reut_a1011)2 from ralstonia eutropha jmp134 at 2.05 a resolution
|
| 74 | c3rbtD_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:glutathione transferase o1;
PDBTitle: crystal structure of glutathione s-transferase omega 3 from the2 silkworm bombyx mori
|
| 75 | c2c8uA_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase 28 kda;
PDBTitle: structure of r21q mutant of sh28gst
|
| 76 | c3touB_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione s-transferase protein;
PDBTitle: crystal structure of glutathione transferase (target efi-501058) from2 ralstonia solanacearum gmi1000 with gsh bound
|
| 77 | c1tw9C_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:glutathione s-transferase 2;
PDBTitle: glutathione transferase-2, apo form, from the nematode heligmosomoides2 polygyrus
|
| 78 | c2r4vA_ |
|
not modelled |
100.0 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:chloride intracellular channel protein 2;
PDBTitle: structure of human clic2, crystal form a
|
| 79 | c3ic8D_ |
|
not modelled |
100.0 |
10 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized gst-like proteinprotein;
PDBTitle: the crystal structure of a gst-like protein from pseudomonas syringae2 to 2.4a
|
| 80 | c3ppuB_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione-s-transferase;
PDBTitle: crystal structure of the glutathione-s-transferase xi from2 phanerochaete chrysosporium
|
| 81 | c3fy7B_ |
|
not modelled |
100.0 |
19 |
PDB header:transport protein
Chain: B: PDB Molecule:chloride intracellular channel protein 3;
PDBTitle: crystal structure of homo sapiens clic3
|
| 82 | c2yv9B_ |
|
not modelled |
100.0 |
12 |
PDB header:metal transport
Chain: B: PDB Molecule:chloride intracellular channel exc-4;
PDBTitle: crystal structure of the clic homologue exc-4 from c.2 elegans
|
| 83 | c2yv7A_ |
|
not modelled |
100.0 |
18 |
PDB header:metal transport
Chain: A: PDB Molecule:cg10997-pa;
PDBTitle: crystal structure of the clic homolog from drosophila2 melanogaster
|
| 84 | c1e6bA_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: crystal structure of a zeta class glutathione s-transferase2 from arabidopsis thaliana
|
| 85 | c3m1gC_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:putative glutathione s-transferase;
PDBTitle: the structure of a putative glutathione s-transferase from2 corynebacterium glutamicum
|
| 86 | c1z9hB_ |
|
not modelled |
99.9 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:membrane-associated prostaglandin e synthase-2;
PDBTitle: microsomal prostaglandin e synthase type-2
|
| 87 | c2uz8A_ |
|
not modelled |
99.9 |
20 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:eukaryotic translation elongation factor 1
PDBTitle: the crystal structure of p18, human translation elongation2 factor 1 epsilon 1
|
| 88 | d1n2aa1 |
|
not modelled |
99.9 |
100 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 89 | d1f2ea1 |
|
not modelled |
99.9 |
32 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 90 | d1pmta1 |
|
not modelled |
99.8 |
48 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 91 | d1n2aa2 |
|
not modelled |
99.8 |
100 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 92 | d1f2ea2 |
|
not modelled |
99.8 |
58 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 93 | d1pmta2 |
|
not modelled |
99.8 |
63 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 94 | c2hsmA_ |
|
not modelled |
99.8 |
14 |
PDB header:ligase/rna binding protein
Chain: A: PDB Molecule:glutamyl-trna synthetase, cytoplasmic;
PDBTitle: structural basis of yeast aminoacyl-trna synthetase complex2 formation revealed by crystal structures of two binary sub-3 complexes
|
| 95 | d1jlwa2 |
|
not modelled |
99.8 |
28 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 96 | d1v2aa2 |
|
not modelled |
99.8 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 97 | d1pn9a2 |
|
not modelled |
99.7 |
30 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 98 | d1axda1 |
|
not modelled |
99.7 |
22 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 99 | d3gtub1 |
|
not modelled |
99.7 |
16 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 100 | d1aw9a1 |
|
not modelled |
99.7 |
23 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 101 | d2c4ja1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 102 | d1b4pa1 |
|
not modelled |
99.7 |
17 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 103 | d1xw6a1 |
|
not modelled |
99.7 |
17 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 104 | d4gtua1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 105 | d1gsua1 |
|
not modelled |
99.7 |
19 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 106 | d2fhea1 |
|
not modelled |
99.7 |
14 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 107 | d2gsta1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 108 | d1jlva1 |
|
not modelled |
99.7 |
16 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 109 | d1ljra2 |
|
not modelled |
99.7 |
29 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 110 | d1nhya1 |
|
not modelled |
99.7 |
11 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 111 | d1e6ba2 |
|
not modelled |
99.7 |
29 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 112 | d1jlva2 |
|
not modelled |
99.7 |
29 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 113 | d1duga1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 114 | d1gnea1 |
|
not modelled |
99.7 |
14 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 115 | d2gsra1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 116 | d1lbka1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 117 | d1gnwa2 |
|
not modelled |
99.7 |
32 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 118 | d2a2ra1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 119 | d1gnwa1 |
|
not modelled |
99.7 |
15 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 120 | d1fw1a1 |
|
not modelled |
99.7 |
16 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |