| 1 |
|
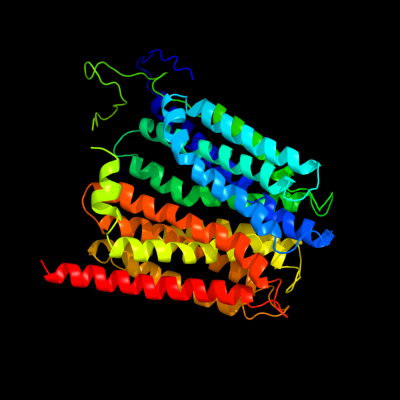
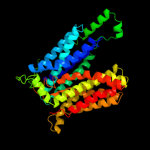
PDB 1pw4 chain A
Region: 21 - 453
Aligned: 419
Modelled: 433
Confidence: 100.0%
Identity: 16%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
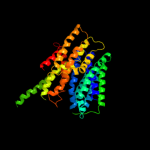
PDB 1pv7 chain A
Region: 36 - 453
Aligned: 408
Modelled: 418
Confidence: 100.0%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
PDB 2gfp chain A
Region: 43 - 434
Aligned: 369
Modelled: 376
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
|
PDB 3o7p chain A
Region: 46 - 444
Aligned: 387
Modelled: 399
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 45 - 439
Aligned: 395
Modelled: 395
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 2g9p chain A
Region: 96 - 109
Aligned: 14
Modelled: 14
Confidence: 12.7%
Identity: 21%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 7 |
|
PDB 3pro chain C domain 1
Region: 64 - 105
Aligned: 42
Modelled: 42
Confidence: 11.4%
Identity: 17%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2
| 8 |
|
PDB 2rdd chain B
Region: 428 - 453
Aligned: 26
Modelled: 26
Confidence: 7.2%
Identity: 15%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 9 |
|
PDB 2axt chain I domain 1
Region: 424 - 453
Aligned: 30
Modelled: 30
Confidence: 6.7%
Identity: 13%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein I, PsbI
Family: PsbI-like
Phyre2
| 10 |
|
PDB 3b9y chain A
Region: 326 - 453
Aligned: 125
Modelled: 128
Confidence: 6.1%
Identity: 8%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 11 |
|
PDB 3prq chain T
Region: 426 - 452
Aligned: 27
Modelled: 27
Confidence: 5.8%
Identity: 11%
PDB header:photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
Phyre2
| 12 |
|
PDB 3bz2 chain T
Region: 426 - 452
Aligned: 27
Modelled: 27
Confidence: 5.8%
Identity: 11%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
Phyre2
| 13 |
|
PDB 3bz1 chain T
Region: 426 - 452
Aligned: 27
Modelled: 27
Confidence: 5.8%
Identity: 11%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
Phyre2