| 1 |
|
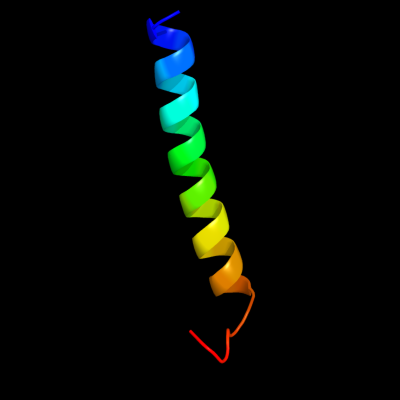
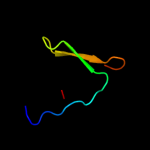
PDB 1szi chain A
Region: 57 - 88
Aligned: 32
Modelled: 32
Confidence: 63.0%
Identity: 22%
Fold: Four-helical up-and-down bundle
Superfamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domain
Family: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domain
Phyre2
| 2 |
|
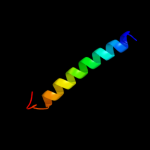
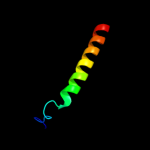
PDB 2cj9 chain A
Region: 22 - 89
Aligned: 44
Modelled: 48
Confidence: 12.5%
Identity: 30%
PDB header:ligase
Chain: A: PDB Molecule:seryl-trna synthetase;
PDBTitle: crystal structure of methanosarcina barkeri seryl-trna2 synthetase complexed with an analog of seryladenylate
Phyre2
| 3 |
|
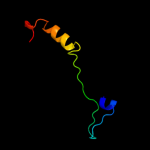
PDB 1tdp chain A
Region: 71 - 83
Aligned: 13
Modelled: 13
Confidence: 12.5%
Identity: 31%
Fold: Bromodomain-like
Superfamily: Bacteriocin immunity protein-like
Family: Carnobacteriocin B2 immunity protein
Phyre2
| 4 |
|
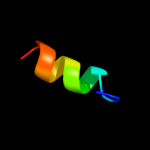
PDB 3mko chain A
Region: 58 - 88
Aligned: 22
Modelled: 30
Confidence: 9.3%
Identity: 23%
PDB header:viral protein
Chain: A: PDB Molecule:glycoprotein c;
PDBTitle: crystal structure of the lymphocytic choriomeningitis virus membrane2 fusion glycoprotein gp2 in its postfusion conformation
Phyre2
| 5 |
|
PDB 2qnl chain A
Region: 62 - 88
Aligned: 27
Modelled: 27
Confidence: 9.1%
Identity: 22%
PDB header:signaling protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative dna damage-inducible protein2 (chu_0679) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
Phyre2
| 6 |
|
PDB 3cs5 chain B
Region: 71 - 83
Aligned: 13
Modelled: 13
Confidence: 6.3%
Identity: 31%
PDB header:photosynthesis
Chain: B: PDB Molecule:phycobilisome degradation protein nbla;
PDBTitle: nbla protein from synechococcus elongatus pcc 7942
Phyre2
| 7 |
|
PDB 2axt chain H domain 1
Region: 44 - 77
Aligned: 34
Modelled: 34
Confidence: 5.6%
Identity: 21%
Fold: Single transmembrane helix
Superfamily: Photosystem II 10 kDa phosphoprotein PsbH
Family: PsbH-like
Phyre2