1 c1klfP_
100.0
13
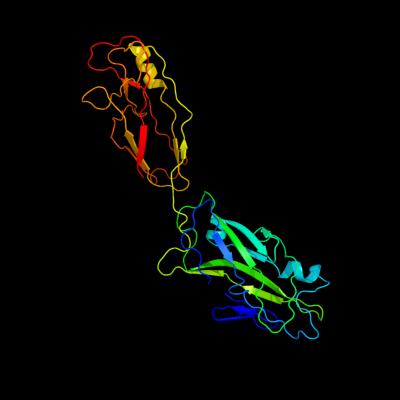
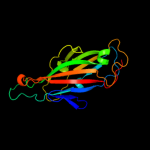
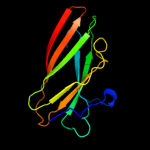
PDB header: chaperone/adhesin complexChain: P: PDB Molecule: fimh protein;PDBTitle: fimh adhesin-fimc chaperone complex with d-mannose
2 c3bfwA_
99.9
14
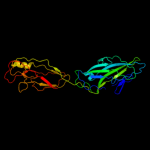
PDB header: structural protein/structural proteinChain: A: PDB Molecule: protein fimg;PDBTitle: crystal structure of truncated fimg (fimgt) in complex with the donor2 strand peptide of fimf (dsf)
3 c3jwnK_
99.8
18
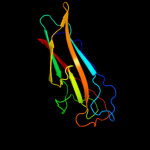
PDB header: protein binding/cell adhesionChain: K: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
4 c3jwnL_
99.8
18
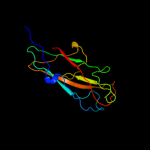
PDB header: protein binding/cell adhesionChain: L: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
5 c3jwnE_
99.8
18
PDB header: protein binding/cell adhesionChain: E: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
6 c3jwnF_
99.8
18
PDB header: protein binding/cell adhesionChain: F: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
7 d1ze3h1
99.8
12
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits8 c2w07B_
99.8
18
PDB header: cell adhesionChain: B: PDB Molecule: minor pilin subunit papf;PDBTitle: structural determinants of polymerization reactivity of the2 p pilus adaptor subunit papf
9 c2jtyA_
99.8
18
PDB header: structural proteinChain: A: PDB Molecule: type-1 fimbrial protein, a chain;PDBTitle: self-complemented variant of fima, the main subunit of type 1 pilus
10 c2jmrA_
99.8
18
PDB header: cell adhesionChain: A: PDB Molecule: fimf;PDBTitle: nmr structure of the e. coli type 1 pilus subunit fimf
11 d2j2zb1
99.8
11
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits12 d1pdkb_
99.8
19
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits13 d2uy6b1
99.8
16
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits14 c3bwuF_
99.8
16
PDB header: chaperone, structural, membrane proteinChain: F: PDB Molecule: protein fimf;PDBTitle: crystal structure of the ternary complex of fimd (n-terminal domain,2 fimdn) with fimc and the n-terminally truncated pilus subunit fimf3 (fimft)
15 d1n12a_
99.6
17
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits16 c1w3gA_
32.0
18
PDB header: toxin/lectinChain: A: PDB Molecule: hemolytic lectin from laetiporus sulphureus;PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
17 c2wmpB_
26.1
12
PDB header: chaperoneChain: B: PDB Molecule: papg protein;PDBTitle: structure of the e. coli chaperone papd in complex with the pilin2 domain of the papgii adhesin
18 d1uwfa1
22.1
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits19 d2jnaa1
11.8
18
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like20 d1p4ua_
11.0
11
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like21 d1na8a_
not modelled
9.6
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like22 d1gywb_
not modelled
6.5
7
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like23 c3l48B_
not modelled
6.2
18
PDB header: transport proteinChain: B: PDB Molecule: outer membrane usher protein papc;PDBTitle: crystal structure of the c-terminal domain of the papc usher
24 d1gyva_
not modelled
6.1
5
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like25 c3ff7B_
not modelled
6.0
33
PDB header: cell adhesion/immunue systemChain: B: PDB Molecule: epithelial cadherin;PDBTitle: structure of nk cell receptor klrg1 bound to e-cadherin