| 1 |
|
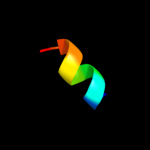
PDB 1jb0 chain M
Region: 120 - 130
Aligned: 11
Modelled: 11
Confidence: 22.1%
Identity: 55%
Fold: Single transmembrane helix
Superfamily: Subunit XII of photosystem I reaction centre, PsaM
Family: Subunit XII of photosystem I reaction centre, PsaM
Phyre2
| 2 |
|
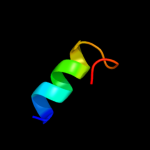
PDB 1jb0 chain M
Region: 120 - 130
Aligned: 11
Modelled: 11
Confidence: 22.1%
Identity: 55%
PDB header:photosynthesis
Chain: M: PDB Molecule:photosystem 1 reaction centre subunit xii;
PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
Phyre2
| 3 |
|
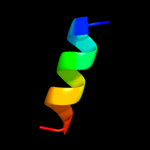
PDB 1jss chain A
Region: 55 - 81
Aligned: 27
Modelled: 27
Confidence: 11.0%
Identity: 22%
Fold: TBP-like
Superfamily: Bet v1-like
Family: STAR domain
Phyre2
| 4 |
|
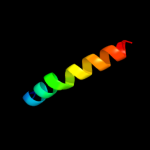
PDB 1jss chain B
Region: 55 - 81
Aligned: 27
Modelled: 27
Confidence: 11.0%
Identity: 22%
PDB header:lipid binding protein
Chain: B: PDB Molecule:cholesterol-regulated start protein 4;
PDBTitle: crystal structure of the mus musculus cholesterol-regulated2 start protein 4 (stard4).
Phyre2
| 5 |
|
PDB 1p0l chain A
Region: 71 - 79
Aligned: 9
Modelled: 9
Confidence: 9.9%
Identity: 33%
PDB header:ribosome
Chain: A: PDB Molecule:19-mer peptide from 50s ribosomal protein l1;
PDBTitle: hp (2-20) substitution gln to trp modification in sds-d252 micelles
Phyre2
| 6 |
|
PDB 2l35 chain B
Region: 125 - 138
Aligned: 14
Modelled: 14
Confidence: 8.5%
Identity: 36%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 7 |
|
PDB 1p0o chain A
Region: 71 - 79
Aligned: 9
Modelled: 9
Confidence: 8.5%
Identity: 33%
PDB header:ribosome
Chain: A: PDB Molecule:19-mer peptide from 50s ribosomal protein l1;
PDBTitle: hp (2-20) substitution of trp for gln and asp at position2 17 and 19 modification in sds-d25 micelles
Phyre2
| 8 |
|
PDB 2jd3 chain B
Region: 127 - 142
Aligned: 16
Modelled: 16
Confidence: 8.3%
Identity: 31%
PDB header:dna binding protein
Chain: B: PDB Molecule:stbb protein;
PDBTitle: parr from plasmid pb171
Phyre2
| 9 |
|
PDB 2l34 chain B
Region: 126 - 138
Aligned: 13
Modelled: 13
Confidence: 7.9%
Identity: 38%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 10 |
|
PDB 2l34 chain A
Region: 126 - 138
Aligned: 13
Modelled: 13
Confidence: 7.9%
Identity: 38%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 11 |
|
PDB 1ln1 chain A
Region: 55 - 82
Aligned: 28
Modelled: 28
Confidence: 6.3%
Identity: 18%
Fold: TBP-like
Superfamily: Bet v1-like
Family: STAR domain
Phyre2
| 12 |
|
PDB 2zt9 chain E
Region: 59 - 69
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 45%
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
Phyre2
| 13 |
|
PDB 1t0a chain A
Region: 117 - 153
Aligned: 37
Modelled: 37
Confidence: 5.5%
Identity: 27%
Fold: Bacillus chorismate mutase-like
Superfamily: IpsF-like
Family: IpsF-like
Phyre2
| 14 |
|
PDB 3dby chain N
Region: 124 - 154
Aligned: 31
Modelled: 31
Confidence: 5.4%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: N: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein from bacillus cereus2 g9241 (csap target)
Phyre2