| 1 |
|
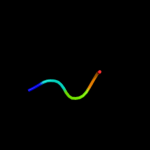
PDB 2k1a chain A
Region: 56 - 84
Aligned: 29
Modelled: 29
Confidence: 39.0%
Identity: 24%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 2 |
|
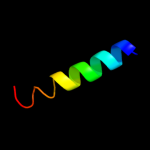
PDB 2knc chain A
Region: 56 - 84
Aligned: 29
Modelled: 29
Confidence: 38.5%
Identity: 24%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 3 |
|
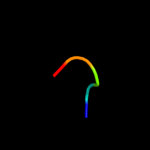
PDB 2bbj chain B
Region: 66 - 84
Aligned: 19
Modelled: 19
Confidence: 14.4%
Identity: 16%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 4 |
|
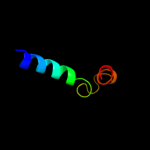
PDB 2jpn chain A
Region: 135 - 142
Aligned: 8
Modelled: 8
Confidence: 13.5%
Identity: 50%
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent dna helicase uvsw;
PDBTitle: solution structure of t4 bacteriophage helicase uvsw.1
Phyre2
| 5 |
|
PDB 2lcy chain A
Region: 129 - 133
Aligned: 5
Modelled: 5
Confidence: 13.5%
Identity: 60%
PDB header:viral protein
Chain: A: PDB Molecule:virion spike glycoprotein;
PDBTitle: nmr structure of the complete internal fusion loop from ebolavirus gp22 at ph 5.5
Phyre2
| 6 |
|
PDB 2iub chain A domain 2
Region: 64 - 84
Aligned: 21
Modelled: 21
Confidence: 12.9%
Identity: 14%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 7 |
|
PDB 3csy chain J
Region: 129 - 133
Aligned: 5
Modelled: 5
Confidence: 8.6%
Identity: 60%
PDB header:immune system/viral protein
Chain: J: PDB Molecule:envelope glycoprotein gp2;
PDBTitle: crystal structure of the trimeric prefusion ebola virus glycoprotein2 in complex with a neutralizing antibody from a human survivor
Phyre2
| 8 |
|
PDB 3s88 chain J
Region: 129 - 133
Aligned: 5
Modelled: 5
Confidence: 7.9%
Identity: 60%
PDB header:immune system/viral protein
Chain: J: PDB Molecule:envelope glycoprotein;
PDBTitle: crystal structure of sudan ebolavirus glycoprotein (strain gulu) bound2 to 16f6
Phyre2
| 9 |
|
PDB 1wix chain A
Region: 112 - 144
Aligned: 26
Modelled: 33
Confidence: 6.6%
Identity: 27%
Fold: CH domain-like
Superfamily: Hook domain
Family: Hook domain
Phyre2
| 10 |
|
PDB 3ehb chain B domain 2
Region: 51 - 103
Aligned: 53
Modelled: 53
Confidence: 6.0%
Identity: 23%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2