| 1 |
|
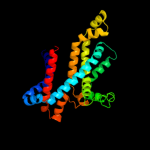
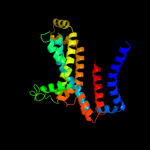
PDB 3fh6 chain F
Region: 9 - 292
Aligned: 281
Modelled: 284
Confidence: 100.0%
Identity: 27%
PDB header:transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
Phyre2
| 2 |
|
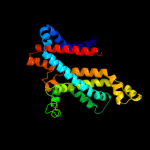
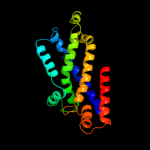
PDB 2r6g chain F
Region: 44 - 282
Aligned: 236
Modelled: 239
Confidence: 100.0%
Identity: 29%
PDB header:hydrolase/transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: the crystal structure of the e. coli maltose transporter
Phyre2
| 3 |
|
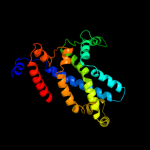
PDB 2r6g chain G domain 1
Region: 9 - 290
Aligned: 264
Modelled: 264
Confidence: 100.0%
Identity: 24%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 4 |
|
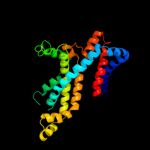
PDB 2onk chain C domain 1
Region: 11 - 288
Aligned: 250
Modelled: 251
Confidence: 100.0%
Identity: 18%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 5 |
|
PDB 2onk chain C
Region: 11 - 288
Aligned: 250
Modelled: 251
Confidence: 100.0%
Identity: 18%
PDB header:membrane protein
Chain: C: PDB Molecule:molybdate/tungstate abc transporter, permease
PDBTitle: abc transporter modbc in complex with its binding protein2 moda
Phyre2
| 6 |
|
PDB 2r6g chain F domain 2
Region: 57 - 282
Aligned: 223
Modelled: 226
Confidence: 100.0%
Identity: 30%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 7 |
|
PDB 3d31 chain C domain 1
Region: 15 - 284
Aligned: 245
Modelled: 246
Confidence: 100.0%
Identity: 20%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 8 |
|
PDB 3d31 chain D
Region: 15 - 284
Aligned: 245
Modelled: 246
Confidence: 100.0%
Identity: 20%
PDB header:transport protein
Chain: D: PDB Molecule:sulfate/molybdate abc transporter, permease
PDBTitle: modbc from methanosarcina acetivorans
Phyre2
| 9 |
|
PDB 3dhw chain A domain 1
Region: 69 - 282
Aligned: 197
Modelled: 199
Confidence: 99.9%
Identity: 18%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 10 |
|
PDB 2r6g chain F domain 1
Region: 9 - 69
Aligned: 56
Modelled: 61
Confidence: 95.2%
Identity: 20%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 11 |
|
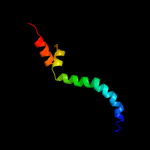
PDB 2hx6 chain A
Region: 180 - 205
Aligned: 26
Modelled: 26
Confidence: 29.7%
Identity: 19%
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease;
PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
Phyre2
| 12 |
|
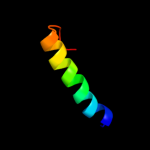
PDB 1umq chain A
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 24.1%
Identity: 19%
PDB header:dna-binding protein
Chain: A: PDB Molecule:photosynthetic apparatus regulatory protein;
PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
Phyre2
| 13 |
|
PDB 1umq chain A
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 24.1%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 14 |
|
PDB 1fip chain A
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 22.4%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 15 |
|
PDB 1eto chain B
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 19.0%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 16 |
|
PDB 1ntc chain A
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 18.0%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 17 |
|
PDB 1etx chain A
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 17.6%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 18 |
|
PDB 1g2h chain A
Region: 181 - 211
Aligned: 30
Modelled: 31
Confidence: 17.4%
Identity: 10%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 19 |
|
PDB 3e7l chain D
Region: 181 - 211
Aligned: 31
Modelled: 31
Confidence: 16.8%
Identity: 23%
PDB header:transcription regulator
Chain: D: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
Phyre2
| 20 |
|
PDB 2cw1 chain A
Region: 190 - 203
Aligned: 14
Modelled: 14
Confidence: 8.3%
Identity: 36%
PDB header:de novo protein
Chain: A: PDB Molecule:sn4m;
PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
Phyre2
| 21 |
|
| 22 |
|