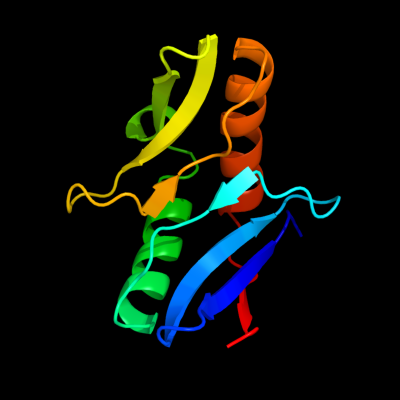
| 1 | c2k49A_
|
|
 |
100.0 |
65 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0339 protein so_3888;
PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
|
| 2 | c2k8eA_
|
|
 |
100.0 |
100 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0339 protein yegp;
PDBTitle: solution nmr structure of protein of unknown function yegp from e.2 coli. ontario center for structural proteomics target ec0640_1_1233 northeast structural genomics consortium target et102.
|
| 3 | d2k49a2
|
|
 |
99.8 |
71 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 4 | d2k8ea1
|
|
 |
99.8 |
100 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 5 | d2k7ia1
|
|
 |
99.8 |
28 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 6 | c2k7iB_
|
|
 |
99.8 |
28 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:upf0339 protein atu0232;
PDBTitle: solution nmr structure of protein atu0232 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att3. ontario center for structural proteomics target atc0223.
|
| 7 | d3bida1
|
|
 |
99.7 |
28 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 8 | d2k8ea2
|
|
 |
99.7 |
100 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 9 | d2k49a1
|
|
 |
99.7 |
60 |
Fold:YegP-like
Superfamily:YegP-like
Family:YegP-like |
| 10 | d1st9a_
|
|
 |
55.7 |
9 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 11 | d1jkea_
|
|
 |
54.0 |
8 |
Fold:DTD-like
Superfamily:DTD-like
Family:DTD-like |
| 12 | c2qz7B_
|
|
 |
52.2 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein sco6318;
PDBTitle: the crystal structure of a homologue of telluride resistance protein2 (terd), sco6318 from streptomyces coelicolor a3(2)
|
| 13 | d1j7ga_
|
|
 |
51.5 |
12 |
Fold:DTD-like
Superfamily:DTD-like
Family:DTD-like |
| 14 | c2l5oA_
|
|
 |
42.6 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative thioredoxin;
PDBTitle: solution structure of a putative thioredoxin from neisseria2 meningitidis
|
| 15 | c3jqxA_
|
|
 |
39.5 |
20 |
PDB header:cell adhesion
Chain: A: PDB Molecule:colh protein;
PDBTitle: crystal structure of clostridium histolyticum colh collagenase2 collagen binding domain 3 at 2.2 angstrom resolution in the presence3 of calcium and cademium
|
| 16 | c3ko7E_
|
|
 |
38.8 |
20 |
PDB header:hydrolase
Chain: E: PDB Molecule:d-tyrosyl-trna(tyr) deacylase;
PDBTitle: dtd from plasmodium falciparum in complex with d-lysine
|
| 17 | c2qngA_
|
|
 |
38.6 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sav2460;
PDBTitle: crytal structure of unknown function protein sav2460
|
| 18 | c2dboA_
|
|
 |
34.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:d-tyrosyl-trna(tyr) deacylase;
PDBTitle: crystal structure of d-tyr-trna(tyr) deacylase from aquifex aeolicus
|
| 19 | d2fy6a1
|
|
 |
33.1 |
6 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 20 | d2b5xa1
|
|
 |
26.9 |
10 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 21 | d1h8la1 |
|
not modelled |
26.6 |
16 |
Fold:Prealbumin-like
Superfamily:Carboxypeptidase regulatory domain-like
Family:Carboxypeptidase regulatory domain |
| 22 | c2b1kA_ |
|
not modelled |
24.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbe;
PDBTitle: crystal structure of e. coli ccmg protein
|
| 23 | c3or5A_ |
|
not modelled |
24.1 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein, thioredoxin family
PDBTitle: crystal structure of thiol:disulfide interchange protein, thioredoxin2 family protein from chlorobium tepidum tls
|
| 24 | c3m86B_ |
|
not modelled |
23.2 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:amoebiasin-2;
PDBTitle: crystal structure of the cysteine protease inhibitor, ehicp2, from2 entamoeba histolytica
|
| 25 | c2f4nA_ |
|
not modelled |
23.0 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein mj1651;
PDBTitle: crystal structure of protein mj1651 from methanococcus2 jannaschii dsm 2661, pfam duf62
|
| 26 | c3cmiA_ |
|
not modelled |
22.8 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peroxiredoxin hyr1;
PDBTitle: crystal structure of glutathione-dependent phospholipid peroxidase2 hyr1 from the yeast saccharomyces cerevisiae
|
| 27 | c3m2oB_ |
|
not modelled |
22.3 |
9 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:glyoxalase/bleomycin resistance protein;
PDBTitle: crystal structure of a putative glyoxalase/bleomycin resistance2 protein from rhodopseudomonas palustris cga009
|
| 28 | c2wgnB_ |
|
not modelled |
22.1 |
21 |
PDB header:hydrolase inhibitor
Chain: B: PDB Molecule:inhibitor of cysteine peptidase compnd 3;
PDBTitle: pseudomonas aeruginosa icp
|
| 29 | d2r6gf1 |
|
not modelled |
21.8 |
8 |
Fold:MalF N-terminal region-like
Superfamily:MalF N-terminal region-like
Family:MalF N-terminal region-like |
| 30 | c2he3A_ |
|
not modelled |
21.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase 2;
PDBTitle: crystal structure of the selenocysteine to cysteine mutant of human2 glutathionine peroxidase 2 (gpx2)
|
| 31 | c3gg4B_ |
|
not modelled |
20.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: the crystal structure of glycerol kinase from yersinia2 pseudotuberculosis
|
| 32 | d1knga_ |
|
not modelled |
19.9 |
6 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 33 | c2kucA_ |
|
not modelled |
19.6 |
3 |
PDB header:isomerase
Chain: A: PDB Molecule:putative disulphide-isomerase;
PDBTitle: solution structure of a putative disulphide-isomerase from bacteroides2 thetaiotaomicron
|
| 34 | c3cxbA_ |
|
not modelled |
19.1 |
7 |
PDB header:signaling protein
Chain: A: PDB Molecule:protein sifa;
PDBTitle: crystal structure of sifa and skip
|
| 35 | d1dfca3 |
|
not modelled |
18.8 |
16 |
Fold:beta-Trefoil
Superfamily:Actin-crosslinking proteins
Family:Fascin |
| 36 | d2hg7a1 |
|
not modelled |
18.8 |
22 |
Fold:gpW/XkdW-like
Superfamily:XkdW-like
Family:XkdW-like |
| 37 | c2hg7A_ |
|
not modelled |
18.8 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phage-like element pbsx protein xkdw;
PDBTitle: solution nmr structure of phage-like element pbsx protein2 xkdw, northeast structural genomics consortium target sr355
|
| 38 | c3g73A_ |
|
not modelled |
18.7 |
26 |
PDB header:transcription/dna
Chain: A: PDB Molecule:forkhead box protein m1;
PDBTitle: structure of the foxm1 dna binding
|
| 39 | c2okvC_ |
|
not modelled |
18.5 |
8 |
PDB header:hydrolase
Chain: C: PDB Molecule:probable d-tyrosyl-trna(tyr) deacylase 1;
PDBTitle: c-myc dna unwinding element binding protein
|
| 40 | c1dfcB_ |
|
not modelled |
18.2 |
11 |
PDB header:structural protein
Chain: B: PDB Molecule:fascin;
PDBTitle: crystal structure of human fascin, an actin-crosslinking protein
|
| 41 | d1k25a3 |
|
not modelled |
17.9 |
18 |
Fold:Penicillin binding protein dimerisation domain
Superfamily:Penicillin binding protein dimerisation domain
Family:Penicillin binding protein dimerisation domain |
| 42 | d2ch5a2 |
|
not modelled |
17.8 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
| 43 | c3razA_ |
|
not modelled |
17.1 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin-related protein;
PDBTitle: the crystal structure of thioredoxin-related protein from neisseria2 meningitidis serogroup b
|
| 44 | c3ibzA_ |
|
not modelled |
17.1 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative tellurium resistant like protein terd;
PDBTitle: crystal structure of putative tellurium resistant like protein (terd)2 from streptomyces coelicolor a3(2)
|
| 45 | c3erwG_ |
|
not modelled |
15.8 |
14 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:sporulation thiol-disulfide oxidoreductase a;
PDBTitle: crystal structure of stoa from bacillus subtilis
|
| 46 | d1ee8a2 |
|
not modelled |
15.2 |
32 |
Fold:N-terminal domain of MutM-like DNA repair proteins
Superfamily:N-terminal domain of MutM-like DNA repair proteins
Family:N-terminal domain of MutM-like DNA repair proteins |
| 47 | c2kxvA_ |
|
not modelled |
15.2 |
21 |
PDB header:unknown function
Chain: A: PDB Molecule:tellurite resistance protein;
PDBTitle: nmr structure and calcium-binding properties of the tellurite2 resistance protein terd from klebsiella pneumoniae
|
| 48 | d1fyhb1 |
|
not modelled |
14.8 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 49 | c1x60A_ |
|
not modelled |
14.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:sporulation-specific n-acetylmuramoyl-l-alanine
PDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
|
| 50 | c2l57A_ |
|
not modelled |
14.5 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of an uncharacterized thioredoin-like protein from2 clostridium perfringens
|
| 51 | d1dfca4 |
|
not modelled |
14.4 |
26 |
Fold:beta-Trefoil
Superfamily:Actin-crosslinking proteins
Family:Fascin |
| 52 | d1z5ye1 |
|
not modelled |
14.2 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 53 | c1k82D_ |
|
not modelled |
14.2 |
18 |
PDB header:hydrolase/dna
Chain: D: PDB Molecule:formamidopyrimidine-dna glycosylase;
PDBTitle: crystal structure of e.coli formamidopyrimidine-dna2 glycosylase (fpg) covalently trapped with dna
|
| 54 | c3eytA_ |
|
not modelled |
14.1 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein spoa0173;
PDBTitle: crystal structure of thioredoxin-like superfamily protein spoa0173
|
| 55 | d1k3xa2 |
|
not modelled |
14.0 |
15 |
Fold:N-terminal domain of MutM-like DNA repair proteins
Superfamily:N-terminal domain of MutM-like DNA repair proteins
Family:N-terminal domain of MutM-like DNA repair proteins |
| 56 | c3f9uA_ |
|
not modelled |
13.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative exported cytochrome c biogenesis-related protein;
PDBTitle: crystal structure of c-terminal domain of putative exported cytochrome2 c biogenesis-related protein from bacteroides fragilis
|
| 57 | d2f8aa1 |
|
not modelled |
13.4 |
10 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 58 | c3nf5A_ |
|
not modelled |
13.4 |
13 |
PDB header:protein transport
Chain: A: PDB Molecule:nucleoporin nup116;
PDBTitle: crystal structure of the c-terminal domain of nuclear pore complex2 component nup116 from candida glabrata
|
| 59 | c2k3iA_ |
|
not modelled |
13.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yiis;
PDBTitle: solution nmr structure of protein yiis from shigella2 flexneri. northeast structural genomics consortium target3 sfr90
|
| 60 | d1y6kr1 |
|
not modelled |
12.7 |
25 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 61 | c3kh7A_ |
|
not modelled |
12.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thiol:disulfide interchange protein dsbe;
PDBTitle: crystal structure of the periplasmic soluble domain of reduced ccmg2 from pseudomonas aeruginosa
|
| 62 | c3ixrA_ |
|
not modelled |
12.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bacterioferritin comigratory protein;
PDBTitle: crystal structure of xylella fastidiosa prxq c47s mutant
|
| 63 | c3hz6A_ |
|
not modelled |
12.2 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:xylulokinase;
PDBTitle: crystal structure of xylulokinase from chromobacterium violaceum
|
| 64 | d2je8a3 |
|
not modelled |
11.6 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 65 | c2p5qA_ |
|
not modelled |
11.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase 5;
PDBTitle: crystal structure of the poplar glutathione peroxidase 5 in2 the reduced form
|
| 66 | c3fcdB_ |
|
not modelled |
11.0 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:lyase;
PDBTitle: crystal structure of a putative glyoxalase from an2 environmental bacteria
|
| 67 | c2rp4C_ |
|
not modelled |
11.0 |
28 |
PDB header:transcription
Chain: C: PDB Molecule:transcription factor p53;
PDBTitle: solution structure of the oligomerization domain in dmp53
|
| 68 | c3fn2A_ |
|
not modelled |
10.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:putative sensor histidine kinase domain;
PDBTitle: crystal structure of a putative sensor histidine kinase domain from2 clostridium symbiosum atcc 14940
|
| 69 | d1wp0a1 |
|
not modelled |
10.9 |
9 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 70 | c2yugA_ |
|
not modelled |
10.7 |
25 |
PDB header:gene regulation
Chain: A: PDB Molecule:protein frg1;
PDBTitle: solution structure of mouse frg1 protein
|
| 71 | d1mpya1 |
|
not modelled |
10.5 |
4 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Extradiol dioxygenases |
| 72 | d1pyya3 |
|
not modelled |
10.3 |
19 |
Fold:Penicillin binding protein dimerisation domain
Superfamily:Penicillin binding protein dimerisation domain
Family:Penicillin binding protein dimerisation domain |
| 73 | c2gzvA_ |
|
not modelled |
10.2 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:prkca-binding protein;
PDBTitle: the cystal structure of the pdz domain of human pick1 (casp target)
|
| 74 | d1k82a2 |
|
not modelled |
9.7 |
18 |
Fold:N-terminal domain of MutM-like DNA repair proteins
Superfamily:N-terminal domain of MutM-like DNA repair proteins
Family:N-terminal domain of MutM-like DNA repair proteins |
| 75 | c2xpoB_ |
|
not modelled |
9.6 |
33 |
PDB header:transcription
Chain: B: PDB Molecule:chromatin structure modulator;
PDBTitle: crystal structure of a spt6-iws1(spn1) complex from2 encephalitozoon cuniculi, form ii
|
| 76 | d1wzla1 |
|
not modelled |
9.6 |
21 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:E-set domains of sugar-utilizing enzymes |
| 77 | c2b7kD_ |
|
not modelled |
9.6 |
18 |
PDB header:metal binding protein
Chain: D: PDB Molecule:sco1 protein;
PDBTitle: crystal structure of yeast sco1
|
| 78 | c2aivA_ |
|
not modelled |
9.6 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:fragment of nucleoporin nup116/nsp116;
PDBTitle: multiple conformations in the ligand-binding site of the2 yeast nuclear pore targeting domain of nup116p
|
| 79 | c3c12A_ |
|
not modelled |
9.4 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:flagellar protein;
PDBTitle: crystal structure of flgd from xanthomonas campestris:2 insights into the hook capping essential for flagellar3 assembly
|
| 80 | c2v1mA_ |
|
not modelled |
9.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase;
PDBTitle: crystal structure of schistosoma mansoni glutathione2 peroxidase
|
| 81 | d1rqpa2 |
|
not modelled |
9.1 |
21 |
Fold:Bacterial fluorinating enzyme, N-terminal domain
Superfamily:Bacterial fluorinating enzyme, N-terminal domain
Family:Bacterial fluorinating enzyme, N-terminal domain |
| 82 | d1aopa2 |
|
not modelled |
8.8 |
15 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 83 | d1t6sa1 |
|
not modelled |
8.7 |
5 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ScpB/YpuH-like |
| 84 | c2p31B_ |
|
not modelled |
8.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione peroxidase 7;
PDBTitle: crystal structure of human glutathione peroxidase 7
|
| 85 | d1rwua_ |
|
not modelled |
8.7 |
16 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:YbeD-like |
| 86 | c1rwuA_ |
|
not modelled |
8.7 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical upf0250 protein ybed;
PDBTitle: solution structure of conserved protein ybed from e. coli
|
| 87 | c2kzrA_ |
|
not modelled |
8.6 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:ubiquitin thioesterase otu1;
PDBTitle: solution nmr structure of ubiquitin thioesterase otu1 (ec 3.1.2.-)2 from mus musculus, northeast structural genomics consortium target3 mmt2a
|
| 88 | d2ho2a1 |
|
not modelled |
8.6 |
16 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 89 | c3dwvB_ |
|
not modelled |
8.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione peroxidase-like protein;
PDBTitle: glutathione peroxidase-type tryparedoxin peroxidase,2 oxidized form
|
| 90 | c2bsdC_ |
|
not modelled |
8.2 |
18 |
PDB header:receptor
Chain: C: PDB Molecule:receptor binding protein;
PDBTitle: structure of lactococcal bacteriophage p2 receptor binding2 protein
|
| 91 | d1tocr1 |
|
not modelled |
8.2 |
18 |
Fold:BPTI-like
Superfamily:BPTI-like
Family:Soft tick anticoagulant proteins |
| 92 | d1ys7a1 |
|
not modelled |
8.1 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:PhoB-like |
| 93 | c3gl3D_ |
|
not modelled |
7.9 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative thiol:disulfide interchange protein
PDBTitle: crystal structure of a putative thiol:disulfide interchange2 protein dsbe from chlorobium tepidum
|
| 94 | c3co7C_ |
|
not modelled |
7.9 |
15 |
PDB header:transcription/dna
Chain: C: PDB Molecule:forkhead box protein o1;
PDBTitle: crystal structure of foxo1 dbd bound to dbe2 dna
|
| 95 | c2cw5B_ |
|
not modelled |
7.7 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:bacterial fluorinating enzyme homolog;
PDBTitle: crystal structure of a conserved hypothetical protein from2 thermus thermophilus hb8
|
| 96 | d1r2za2 |
|
not modelled |
7.7 |
14 |
Fold:N-terminal domain of MutM-like DNA repair proteins
Superfamily:N-terminal domain of MutM-like DNA repair proteins
Family:N-terminal domain of MutM-like DNA repair proteins |
| 97 | d1m7xa1 |
|
not modelled |
7.7 |
29 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:E-set domains of sugar-utilizing enzymes |
| 98 | c3g12A_ |
|
not modelled |
7.6 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:putative lactoylglutathione lyase;
PDBTitle: crystal structure of a putative lactoylglutathione lyase2 from bdellovibrio bacteriovorus
|
| 99 | c1w3gA_ |
|
not modelled |
7.5 |
24 |
PDB header:toxin/lectin
Chain: A: PDB Molecule:hemolytic lectin from laetiporus sulphureus;
PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
|