| 1 |
|
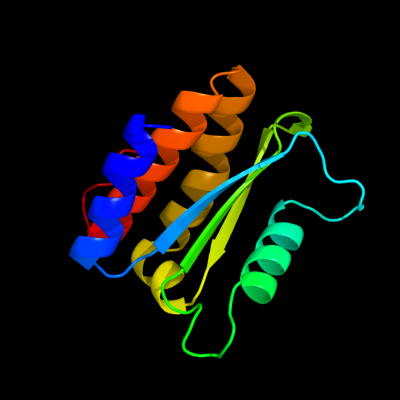
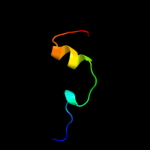
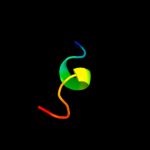
PDB 3ptj chain A
Region: 41 - 162
Aligned: 122
Modelled: 122
Confidence: 98.1%
Identity: 8%
PDB header:hydrolase
Chain: A: PDB Molecule:upf0603 protein at1g54780, chloroplastic;
PDBTitle: structural and functional analysis of arabidopsis thaliana thylakoid2 lumen protein attlp18.3
Phyre2
| 2 |
|
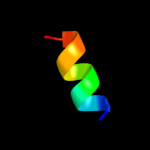
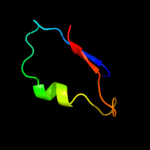
PDB 2kw7 chain A
Region: 32 - 155
Aligned: 123
Modelled: 124
Confidence: 97.9%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: solution nmr structure of the n-terminal domain of protein pg_03612 from p.gingivalis, northeast structural genomics consortium target3 pgr37a
Phyre2
| 3 |
|
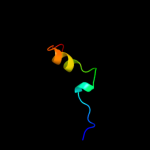
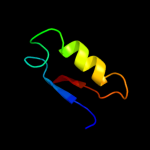
PDB 2kpt chain A
Region: 40 - 161
Aligned: 111
Modelled: 119
Confidence: 62.0%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative secreted protein;
PDBTitle: solution nmr structure of the n-terminal domain of cg24962 protein from corynebacterium glutamicum. northeast3 structural genomics consortium target cgr26a
Phyre2
| 4 |
|
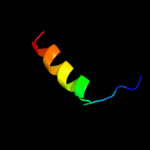
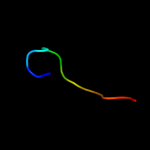
PDB 2y69 chain S
Region: 36 - 60
Aligned: 25
Modelled: 25
Confidence: 22.8%
Identity: 32%
PDB header:electron transport
Chain: S: PDB Molecule:cytochrome c oxidase subunit 5b;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 5 |
|
PDB 1q90 chain N
Region: 187 - 197
Aligned: 11
Modelled: 11
Confidence: 18.4%
Identity: 64%
Fold: Single transmembrane helix
Superfamily: PetN subunit of the cytochrome b6f complex
Family: PetN subunit of the cytochrome b6f complex
Phyre2
| 6 |
|
PDB 1v54 chain F
Region: 35 - 60
Aligned: 26
Modelled: 26
Confidence: 15.8%
Identity: 31%
Fold: Rubredoxin-like
Superfamily: Rubredoxin-like
Family: Cytochrome c oxidase Subunit F
Phyre2
| 7 |
|
PDB 1ppr chain M domain 2
Region: 36 - 55
Aligned: 20
Modelled: 20
Confidence: 13.2%
Identity: 25%
Fold: Peridinin-chlorophyll protein
Superfamily: Peridinin-chlorophyll protein
Family: Peridinin-chlorophyll protein
Phyre2
| 8 |
|
PDB 3ism chain C
Region: 160 - 170
Aligned: 11
Modelled: 11
Confidence: 12.3%
Identity: 64%
PDB header:hydrolase inhibitor/hydrolase
Chain: C: PDB Molecule:cg4930;
PDBTitle: crystal structure of the endog/endogi complex: mechanism of endog2 inhibition
Phyre2
| 9 |
|
PDB 3mpo chain D
Region: 27 - 62
Aligned: 32
Modelled: 36
Confidence: 11.7%
Identity: 22%
PDB header:hydrolase
Chain: D: PDB Molecule:predicted hydrolase of the had superfamily;
PDBTitle: the crystal structure of a hydrolase from lactobacillus brevis
Phyre2
| 10 |
|
PDB 1wr8 chain A
Region: 27 - 62
Aligned: 35
Modelled: 36
Confidence: 10.3%
Identity: 23%
Fold: HAD-like
Superfamily: HAD-like
Family: Predicted hydrolases Cof
Phyre2
| 11 |
|
PDB 3svi chain A
Region: 35 - 44
Aligned: 10
Modelled: 10
Confidence: 8.4%
Identity: 60%
PDB header:signaling protein
Chain: A: PDB Molecule:type iii effector hopab2;
PDBTitle: structure of the pto-binding domain of hoppmal generated by limited2 thermolysin digestion
Phyre2
| 12 |
|
PDB 2axt chain M domain 1
Region: 182 - 200
Aligned: 18
Modelled: 19
Confidence: 8.3%
Identity: 50%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein M, PsbM
Family: PsbM-like
Phyre2
| 13 |
|
PDB 1dwn chain A
Region: 71 - 77
Aligned: 7
Modelled: 7
Confidence: 6.7%
Identity: 100%
Fold: RNA bacteriophage capsid protein
Superfamily: RNA bacteriophage capsid protein
Family: RNA bacteriophage capsid protein
Phyre2
| 14 |
|
PDB 2wnm chain A
Region: 229 - 241
Aligned: 13
Modelled: 13
Confidence: 6.4%
Identity: 38%
PDB header:hydrolase
Chain: A: PDB Molecule:gene 2;
PDBTitle: solution structure of gp2
Phyre2
| 15 |
|
PDB 2apj chain A domain 1
Region: 139 - 147
Aligned: 9
Modelled: 9
Confidence: 6.2%
Identity: 56%
Fold: Flavodoxin-like
Superfamily: SGNH hydrolase
Family: Putative acetylxylan esterase-like
Phyre2
| 16 |
|
PDB 1zmb chain A domain 1
Region: 139 - 147
Aligned: 9
Modelled: 9
Confidence: 6.2%
Identity: 44%
Fold: Flavodoxin-like
Superfamily: SGNH hydrolase
Family: Putative acetylxylan esterase-like
Phyre2
| 17 |
|
PDB 1gkp chain A domain 1
Region: 107 - 139
Aligned: 33
Modelled: 33
Confidence: 5.8%
Identity: 18%
Fold: Composite domain of metallo-dependent hydrolases
Superfamily: Composite domain of metallo-dependent hydrolases
Family: Hydantoinase (dihydropyrimidinase)
Phyre2
| 18 |
|
PDB 2r5v chain A
Region: 111 - 177
Aligned: 53
Modelled: 53
Confidence: 5.6%
Identity: 23%
PDB header:oxidoreductase
Chain: A: PDB Molecule:pcza361.1;
PDBTitle: hydroxymandelate synthase crystal structure
Phyre2
| 19 |
|
PDB 1f20 chain A
Region: 55 - 104
Aligned: 36
Modelled: 50
Confidence: 5.3%
Identity: 33%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitric-oxide synthase;
PDBTitle: crystal structure of rat neuronal nitric-oxide synthase fad/nadp+2 domain at 1.9a resolution.
Phyre2
| 20 |
|
PDB 2kne chain B
Region: 139 - 155
Aligned: 13
Modelled: 17
Confidence: 5.3%
Identity: 54%
PDB header:metal transport
Chain: B: PDB Molecule:atpase, ca++ transporting, plasma membrane 4;
PDBTitle: calmodulin wraps around its binding domain in the plasma2 membrane ca2+ pump anchored by a novel 18-1 motif
Phyre2