| 1 |
|
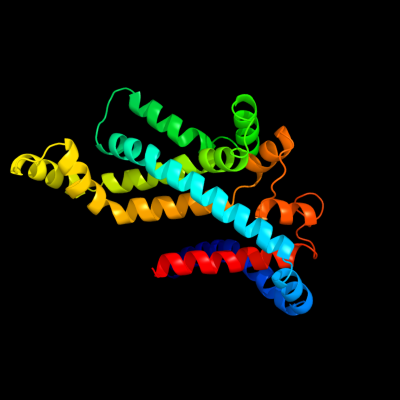
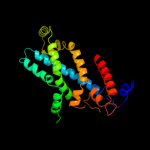
PDB 2onk chain C
Region: 22 - 287
Aligned: 235
Modelled: 242
Confidence: 99.9%
Identity: 16%
PDB header:membrane protein
Chain: C: PDB Molecule:molybdate/tungstate abc transporter, permease
PDBTitle: abc transporter modbc in complex with its binding protein2 moda
Phyre2
| 2 |
|
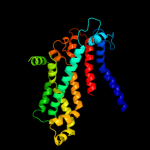
PDB 2onk chain C domain 1
Region: 22 - 287
Aligned: 235
Modelled: 242
Confidence: 99.9%
Identity: 16%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 3 |
|
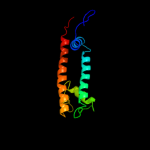
PDB 3dhw chain A domain 1
Region: 89 - 285
Aligned: 189
Modelled: 197
Confidence: 99.9%
Identity: 21%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 4 |
|
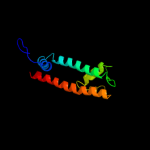
PDB 2r6g chain F domain 2
Region: 71 - 282
Aligned: 208
Modelled: 212
Confidence: 99.9%
Identity: 15%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 5 |
|
PDB 3d31 chain C domain 1
Region: 89 - 283
Aligned: 188
Modelled: 194
Confidence: 99.9%
Identity: 18%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 6 |
|
PDB 3d31 chain D
Region: 89 - 283
Aligned: 188
Modelled: 194
Confidence: 99.9%
Identity: 18%
PDB header:transport protein
Chain: D: PDB Molecule:sulfate/molybdate abc transporter, permease
PDBTitle: modbc from methanosarcina acetivorans
Phyre2
| 7 |
|
PDB 3fh6 chain F
Region: 16 - 296
Aligned: 275
Modelled: 281
Confidence: 99.9%
Identity: 15%
PDB header:transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
Phyre2
| 8 |
|
PDB 2r6g chain F
Region: 66 - 283
Aligned: 213
Modelled: 218
Confidence: 99.9%
Identity: 14%
PDB header:hydrolase/transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: the crystal structure of the e. coli maltose transporter
Phyre2
| 9 |
|
PDB 2r6g chain G domain 1
Region: 24 - 285
Aligned: 259
Modelled: 262
Confidence: 99.8%
Identity: 12%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 10 |
|
PDB 1p7b chain B
Region: 4 - 129
Aligned: 119
Modelled: 126
Confidence: 73.3%
Identity: 8%
PDB header:metal transport
Chain: B: PDB Molecule:integral membrane channel and cytosolic domains;
PDBTitle: crystal structure of an inward rectifier potassium channel
Phyre2
| 11 |
|
PDB 1p7b chain A domain 2
Region: 4 - 125
Aligned: 115
Modelled: 122
Confidence: 47.0%
Identity: 8%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 12 |
|
PDB 2jwa chain A
Region: 96 - 129
Aligned: 34
Modelled: 34
Confidence: 24.8%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 13 |
|
PDB 2voy chain B
Region: 7 - 41
Aligned: 35
Modelled: 35
Confidence: 9.2%
Identity: 9%
PDB header:hydrolase
Chain: B: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 14 |
|
PDB 2lhu chain A
Region: 177 - 206
Aligned: 30
Modelled: 30
Confidence: 7.6%
Identity: 17%
PDB header:structural protein
Chain: A: PDB Molecule:mybpc3 protein;
PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
Phyre2
| 15 |
|
PDB 1umq chain A
Region: 175 - 206
Aligned: 32
Modelled: 32
Confidence: 7.2%
Identity: 6%
PDB header:dna-binding protein
Chain: A: PDB Molecule:photosynthetic apparatus regulatory protein;
PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
Phyre2
| 16 |
|
PDB 1umq chain A
Region: 175 - 206
Aligned: 32
Modelled: 32
Confidence: 7.2%
Identity: 6%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 17 |
|
PDB 1ntc chain A
Region: 175 - 207
Aligned: 33
Modelled: 33
Confidence: 6.3%
Identity: 18%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 18 |
|
PDB 1fip chain A
Region: 175 - 207
Aligned: 33
Modelled: 33
Confidence: 5.7%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 19 |
|
PDB 1eto chain B
Region: 175 - 207
Aligned: 33
Modelled: 33
Confidence: 5.7%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2