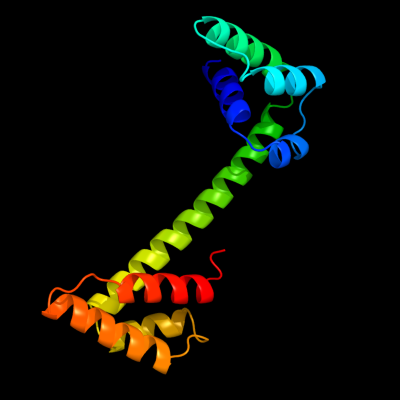
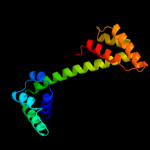
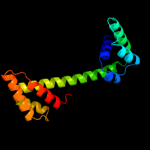
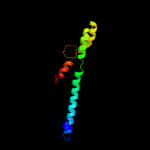
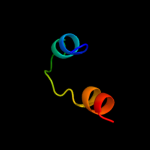
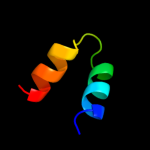
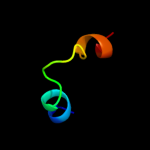
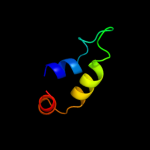
1 d2p0ta1
100.0
41
Fold: PSPTO4464-likeSuperfamily: PSPTO4464-likeFamily: PSPTO4464-like2 c2p0tA_
100.0
41
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0307 protein pspto_4464;PDBTitle: structural genomics, the crystal structure of a conserved putative2 protein from pseudomonas syringae pv. tomato str. dc3000
3 c1nafA_
60.9
13
PDB header: signaling protein, membrane proteinChain: A: PDB Molecule: adp-ribosylation factor binding protein gga1;PDBTitle: crystal structure of the human gga1 gat domain
4 c2kpqA_
50.1
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of agrobacterium tumefaciens protein atu1219:2 northeast structural genomics consortium target att14
5 c2kp6A_
42.9
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein cv0237 from2 chromobacterium violaceum. northeast structural genomics3 consortium (nesg) target cvt1
6 d1tp6a_
40.8
20
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: PA1314-like7 c3opcB_
37.1
18
PDB header: chaperoneChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of flgn chaperone from bordetella pertussis
8 c3c1dA_
26.2
13
PDB header: recombination, dna binding proteinChain: A: PDB Molecule: regulatory protein recx;PDBTitle: x-ray crystal structure of recx
9 d1oxza_
25.9
13
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain10 c1oxzA_
25.9
13
PDB header: membrane proteinChain: A: PDB Molecule: adp-ribosylation factor binding protein gga1;PDBTitle: crystal structure of the human gga1 gat domain
11 c2bcwC_
22.2
16
PDB header: ribosomeChain: C: PDB Molecule: elongation factor g;PDBTitle: coordinates of the n-terminal domain of ribosomal protein2 l11,c-terminal domain of ribosomal protein l7/l12 and a3 portion of the g' domain of elongation factor g, as fitted4 into cryo-em map of an escherichia coli 70s*ef-5 g*gdp*fusidic acid complex
12 d1yt3a2
20.2
23
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: RNase D C-terminal domains13 d2oc6a1
18.5
36
Fold: Secretion chaperone-likeSuperfamily: YdhG-likeFamily: YdhG-like14 c2dhyA_
14.2
15
PDB header: immune systemChain: A: PDB Molecule: cue domain-containing protein 1;PDBTitle: solution structure of the cue domain in the human cue2 domain containing protein 1 (cuedc1)
15 d2i8da1
13.2
26
Fold: Secretion chaperone-likeSuperfamily: YdhG-likeFamily: YdhG-like16 d1fs2b1
12.9
13
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like17 d1fs1b1
12.5
13
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like18 c3k6qB_
10.5
15
PDB header: ligand binding proteinChain: B: PDB Molecule: putative ligand binding protein;PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
19 d1nexa1
10.1
11
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like20 d2ovra1
9.4
13
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like21 d2izva1
not modelled
9.4
12
Fold: SOCS box-likeSuperfamily: SOCS box-likeFamily: SOCS box-like22 c3oq9C_
not modelled
9.2
9
PDB header: apoptosisChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 6;PDBTitle: structure of the fas/fadd death domain assembly
23 c2kl4A_
not modelled
9.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh2032 protein;PDBTitle: nmr structure of the protein nb7804a
24 d1fkma2
not modelled
8.2
17
Fold: Left-handed superhelixSuperfamily: Ypt/Rab-GAP domain of gyp1pFamily: Ypt/Rab-GAP domain of gyp1p25 c3tj3C_
not modelled
7.9
15
PDB header: protein transportChain: C: PDB Molecule: nuclear pore complex protein nup50;PDBTitle: structure of importin a5 bound to the n-terminus of nup50
26 d1k91a_
not modelled
7.7
43
Fold: P-domain of calnexin/calreticulinSuperfamily: P-domain of calnexin/calreticulinFamily: P-domain of calnexin/calreticulin27 c2rqpA_
not modelled
7.2
22
PDB header: gene regulationChain: A: PDB Molecule: heterochromatin protein 1-binding protein 3;PDBTitle: the solution structure of heterochromatin protein 1-binding2 protein 74 histone h1 like domain
28 d2igsa1
not modelled
6.6
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: PA2222-like29 d1in0a1
not modelled
6.6
18
Fold: Ferredoxin-likeSuperfamily: YajQ-likeFamily: YajQ-like30 c3dhyC_
not modelled
6.4
18
PDB header: hydrolaseChain: C: PDB Molecule: adenosylhomocysteinase;PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
31 d1g8eb_
not modelled
6.4
11
Fold: Flagellar transcriptional activator FlhDSuperfamily: Flagellar transcriptional activator FlhDFamily: Flagellar transcriptional activator FlhD32 d2es9a1
not modelled
6.0
45
Fold: YoaC-likeSuperfamily: YoaC-likeFamily: YoaC-like33 c3gehA_
not modelled
6.0
20
PDB header: hydrolaseChain: A: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
34 d2choa2
not modelled
5.9
27
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain35 c2kqrA_
not modelled
5.8
13
PDB header: ligaseChain: A: PDB Molecule: asparaginyl-trna synthetase, cytoplasmic;PDBTitle: solution structure of the n-terminal domain (residues 1-111) of brugia2 malayi asparaginyl-trna synthetase
36 d1sm2a_
not modelled
5.8
24
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Protein kinases, catalytic subunit37 d2cbia2
not modelled
5.7
20
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain38 d1li4a2
not modelled
5.6
12
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: S-adenosylhomocystein hydrolase39 d1v8ba2
not modelled
5.4
23
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: S-adenosylhomocystein hydrolase40 c2j8pA_
not modelled
5.4
17
PDB header: nuclear proteinChain: A: PDB Molecule: cleavage stimulation factor 64 kda subunit;PDBTitle: nmr structure of c-terminal domain of human cstf-64
41 c1d4fD_
not modelled
5.4
12
PDB header: hydrolaseChain: D: PDB Molecule: s-adenosylhomocysteine hydrolase;PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
42 c3d5lA_
not modelled
5.4
16
PDB header: signaling proteinChain: A: PDB Molecule: regulatory protein recx;PDBTitle: crystal structure of regulatory protein recx
43 d2edua1
not modelled
5.3
10
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: ComEA-like44 d2esna1
not modelled
5.1
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LysR-like transcriptional regulators45 d1g8ea_
not modelled
5.0
11
Fold: Flagellar transcriptional activator FlhDSuperfamily: Flagellar transcriptional activator FlhDFamily: Flagellar transcriptional activator FlhD