| 1 |
|
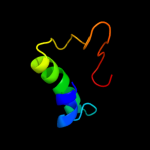
PDB 1iwg chain A domain 8
Region: 208 - 316
Aligned: 106
Modelled: 109
Confidence: 31.5%
Identity: 20%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 2 |
|
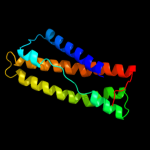
PDB 1ej5 chain A
Region: 181 - 198
Aligned: 18
Modelled: 18
Confidence: 16.6%
Identity: 61%
Fold: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Superfamily: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Family: Wiscott-Aldrich syndrome protein, WASP, C-terminal domain
Phyre2
| 3 |
|
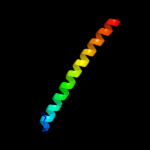
PDB 2itb chain A domain 1
Region: 124 - 141
Aligned: 18
Modelled: 18
Confidence: 14.0%
Identity: 17%
Fold: Ferritin-like
Superfamily: Ferritin-like
Family: MiaE-like
Phyre2
| 4 |
|
PDB 2d6y chain A domain 1
Region: 92 - 138
Aligned: 38
Modelled: 47
Confidence: 9.4%
Identity: 29%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Tetracyclin repressor-like, N-terminal domain
Phyre2
| 5 |
|
PDB 1avo chain A
Region: 24 - 31
Aligned: 8
Modelled: 8
Confidence: 9.1%
Identity: 25%
PDB header:proteasome activator
Chain: A: PDB Molecule:11s regulator;
PDBTitle: proteasome activator reg(alpha)
Phyre2
| 6 |
|
PDB 2o01 chain J
Region: 23 - 39
Aligned: 16
Modelled: 17
Confidence: 8.8%
Identity: 31%
PDB header:photosynthesis
Chain: J: PDB Molecule:photosystem i reaction center subunit ix;
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 7 |
|
PDB 3aqp chain B
Region: 142 - 319
Aligned: 132
Modelled: 133
Confidence: 8.5%
Identity: 18%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 8 |
|
PDB 2nww chain A domain 1
Region: 15 - 98
Aligned: 84
Modelled: 84
Confidence: 7.9%
Identity: 27%
Fold: Proton glutamate symport protein
Superfamily: Proton glutamate symport protein
Family: Proton glutamate symport protein
Phyre2
| 9 |
|
PDB 2k3u chain A
Region: 184 - 200
Aligned: 17
Modelled: 17
Confidence: 6.1%
Identity: 41%
PDB header:immune system
Chain: A: PDB Molecule:chemotaxis inhibitory protein;
PDBTitle: structure of the tyrosine-sulfated c5a receptor n-terminus2 in complex with the immune evasion protein chips.
Phyre2
| 10 |
|
PDB 1r3j chain C
Region: 275 - 319
Aligned: 45
Modelled: 45
Confidence: 5.1%
Identity: 9%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2