| 1 |
|
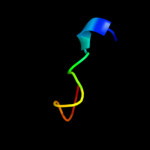
PDB 4a1o chain B
Region: 108 - 117
Aligned: 10
Modelled: 10
Confidence: 20.7%
Identity: 40%
PDB header:transferase-hydrolase
Chain: B: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
Phyre2
| 2 |
|
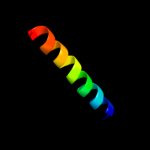
PDB 1zcz chain A
Region: 100 - 117
Aligned: 18
Modelled: 18
Confidence: 18.9%
Identity: 22%
PDB header:transferase/hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of phosphoribosylaminoimidazolecarboxamide2 formyltransferase / imp cyclohydrolase (tm1249) from thermotoga3 maritima at 1.88 a resolution
Phyre2
| 3 |
|
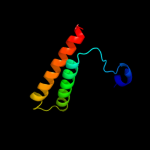
PDB 1thz chain A
Region: 108 - 117
Aligned: 10
Modelled: 10
Confidence: 18.5%
Identity: 40%
PDB header:transferase, hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of avian aicar transformylase in complex2 with a novel inhibitor identified by virtual ligand3 screening
Phyre2
| 4 |
|
PDB 2dyo chain B
Region: 66 - 75
Aligned: 10
Modelled: 10
Confidence: 11.0%
Identity: 60%
PDB header:protein turnover/protein turnover
Chain: B: PDB Molecule:autophagy protein 16;
PDBTitle: the crystal structure of saccharomyces cerevisiae atg5-2 atg16(1-57) complex
Phyre2
| 5 |
|
PDB 3rf1 chain B
Region: 103 - 115
Aligned: 13
Modelled: 13
Confidence: 9.1%
Identity: 31%
PDB header:ligase
Chain: B: PDB Molecule:glycyl-trna synthetase alpha subunit;
PDBTitle: the crystal structure of glycyl-trna synthetase subunit alpha from2 campylobacter jejuni subsp. jejuni nctc 11168
Phyre2
| 6 |
|
PDB 1lir chain A
Region: 67 - 76
Aligned: 10
Modelled: 10
Confidence: 8.1%
Identity: 50%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 7 |
|
PDB 1bzg chain A
Region: 71 - 77
Aligned: 7
Modelled: 7
Confidence: 8.0%
Identity: 57%
PDB header:hormone
Chain: A: PDB Molecule:parathyroid hormone-related protein;
PDBTitle: the solution structure of human parathyroid hormone-related2 protein (1-34) in near-physiological solution, nmr, 303 structures
Phyre2
| 8 |
|
PDB 2axt chain I domain 1
Region: 82 - 107
Aligned: 26
Modelled: 26
Confidence: 7.9%
Identity: 19%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein I, PsbI
Family: PsbI-like
Phyre2
| 9 |
|
PDB 2l2t chain A
Region: 47 - 70
Aligned: 24
Modelled: 24
Confidence: 7.5%
Identity: 21%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 10 |
|
PDB 1gng chain X
Region: 67 - 72
Aligned: 6
Modelled: 6
Confidence: 7.0%
Identity: 83%
PDB header:transferase
Chain: X: PDB Molecule:frattide;
PDBTitle: glycogen synthase kinase-3 beta (gsk3) complex with frattide2 peptide
Phyre2
| 11 |
|
PDB 3skd chain A
Region: 71 - 76
Aligned: 6
Modelled: 6
Confidence: 6.5%
Identity: 50%
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein tthb187;
PDBTitle: crystal structure of the thermus thermophilus cas3 hd domain in the2 presence of ni2+
Phyre2
| 12 |
|
PDB 1y5i chain C domain 1
Region: 4 - 73
Aligned: 70
Modelled: 70
Confidence: 5.7%
Identity: 14%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 13 |
|
PDB 2ogi chain A
Region: 71 - 76
Aligned: 6
Modelled: 6
Confidence: 5.2%
Identity: 50%
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein sag1661;
PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
Phyre2
| 14 |
|
PDB 2rdd chain B
Region: 82 - 104
Aligned: 23
Modelled: 23
Confidence: 5.1%
Identity: 4%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2