| 1 |
|
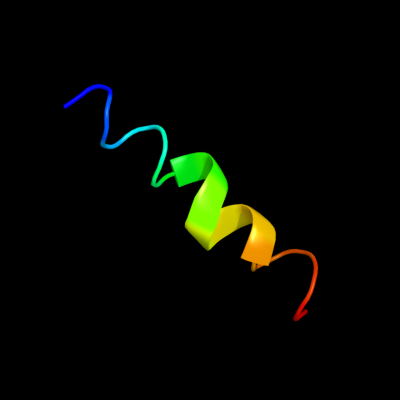
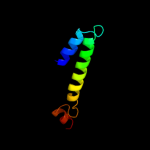
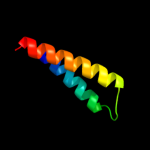
PDB 2wss chain T
Region: 48 - 66
Aligned: 19
Modelled: 19
Confidence: 28.8%
Identity: 26%
PDB header:hydrolase
Chain: T: PDB Molecule:atp synthase subunit b, mitochondrial;
PDBTitle: the structure of the membrane extrinsic region of bovine2 atp synthase
Phyre2
| 2 |
|
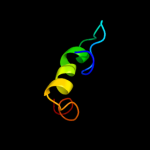
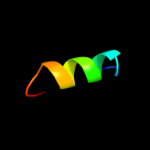
PDB 3d19 chain E
Region: 69 - 115
Aligned: 47
Modelled: 47
Confidence: 25.4%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:conserved metalloprotein;
PDBTitle: crystal structure of a conserved metalloprotein from bacillus cereus
Phyre2
| 3 |
|
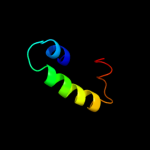
PDB 1d8b chain A
Region: 58 - 87
Aligned: 29
Modelled: 30
Confidence: 15.3%
Identity: 10%
Fold: SAM domain-like
Superfamily: HRDC-like
Family: HRDC domain from helicases
Phyre2
| 4 |
|
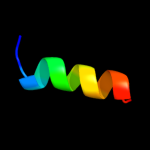
PDB 3dby chain N
Region: 69 - 115
Aligned: 45
Modelled: 47
Confidence: 15.2%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: N: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein from bacillus cereus2 g9241 (csap target)
Phyre2
| 5 |
|
PDB 2lf0 chain A
Region: 52 - 106
Aligned: 55
Modelled: 55
Confidence: 8.7%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yibl;
PDBTitle: solution structure of sf3636, a two-domain unknown function protein2 from shigella flexneri 2a, determined by joint refinement of nmr,3 residual dipolar couplings and small-angle x-ray scatting, nesg4 target sfr339/ocsp target sf3636
Phyre2
| 6 |
|
PDB 2k47 chain A
Region: 89 - 121
Aligned: 33
Modelled: 33
Confidence: 7.0%
Identity: 18%
PDB header:replication
Chain: A: PDB Molecule:phosphoprotein;
PDBTitle: solution structure of the c-terminal n-rna binding domain2 of the vesicular stomatitis virus phosphoprotein
Phyre2
| 7 |
|
PDB 1q2z chain A
Region: 56 - 93
Aligned: 38
Modelled: 38
Confidence: 6.8%
Identity: 18%
Fold: alpha-alpha superhelix
Superfamily: C-terminal domain of Ku80
Family: C-terminal domain of Ku80
Phyre2
| 8 |
|
PDB 2kel chain B
Region: 104 - 118
Aligned: 15
Modelled: 15
Confidence: 6.4%
Identity: 27%
PDB header:transcription repressor
Chain: B: PDB Molecule:uncharacterized protein 56b;
PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
Phyre2
| 9 |
|
PDB 3d19 chain A domain 1
Region: 70 - 115
Aligned: 46
Modelled: 46
Confidence: 5.8%
Identity: 11%
Fold: Bromodomain-like
Superfamily: Bacillus cereus metalloprotein-like
Family: Bacillus cereus metalloprotein-like
Phyre2
| 10 |
|
PDB 1s58 chain A
Region: 66 - 82
Aligned: 17
Modelled: 17
Confidence: 5.5%
Identity: 12%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: ssDNA viruses
Family: Parvoviridae-like VP
Phyre2
| 11 |
|
PDB 2k59 chain B
Region: 8 - 29
Aligned: 22
Modelled: 22
Confidence: 5.4%
Identity: 18%
PDB header:transport protein
Chain: B: PDB Molecule:neuronal acetylcholine receptor subunit beta-2;
PDBTitle: nmr structures of the second transmembrane domain of the2 neuronal acetylcholine receptor beta 2 subunit
Phyre2