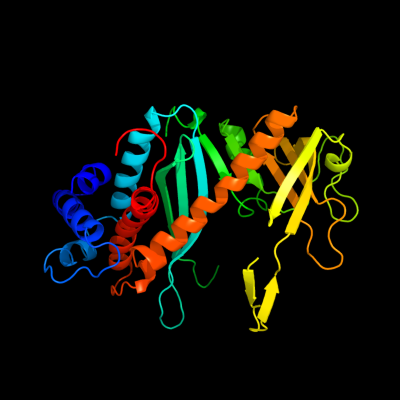
1 d1musa_
99.9
17
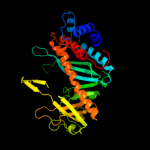
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)2 d1b7ea_
99.9
17
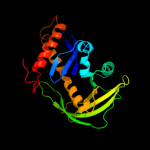
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Transposase inhibitor (Tn5 transposase)3 d3pvia_
28.3
23
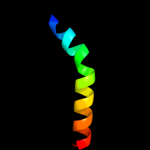
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease PvuII4 c2adcA_
26.8
19
PDB header: rna binding protein/rnaChain: A: PDB Molecule: polypyrimidine tract-binding protein 1;PDBTitle: solution structure of polypyrimidine tract binding protein2 rbd34 complexed with cucucu rna
5 c3hyiA_
19.7
21
PDB header: transcription regulatorChain: A: PDB Molecule: protein duf199/whia;PDBTitle: crystal structure of full-length duf199/whia from thermatoga maritima
6 d2adca1
18.8
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD7 c3hefB_
18.1
15
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
8 c1qm9A_
17.3
15
PDB header: ribonucleoproteinChain: A: PDB Molecule: polypyrimidine tract-binding protein;PDBTitle: nmr, representative structure
9 c3kskA_
15.6
23
PDB header: hydrolaseChain: A: PDB Molecule: type-2 restriction enzyme pvuii;PDBTitle: crystal structure of single chain pvuii
10 c2kviA_
12.9
20
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein 3;PDBTitle: structure of nab3 rrm
11 d2ctsa_
11.8
10
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase12 c2xigA_
10.9
11
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
13 d1ug7a_
10.5
30
Fold: Four-helical up-and-down bundleSuperfamily: Domain from hypothetical 2610208m17rik proteinFamily: Domain from hypothetical 2610208m17rik protein14 c2q9rA_
10.1
13
PDB header: unknown functionChain: A: PDB Molecule: protein of unknown function;PDBTitle: crystal structure of a duf416 family protein (sbal_3149) from2 shewanella baltica os155 at 1.91 a resolution
15 d1x4da1
9.6
7
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD16 d1csha_
9.1
12
Fold: Citrate synthaseSuperfamily: Citrate synthaseFamily: Citrate synthase17 d2qklb1
9.1
48
Fold: Dcp2 domain-likeSuperfamily: Dcp2 domain-likeFamily: Dcp2 box A domain18 d2ezha_
8.9
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain19 d2ezia_
7.8
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain20 c2yy0D_
7.6
26
PDB header: transcriptionChain: D: PDB Molecule: c-myc-binding protein;PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
21 d1ftha_
not modelled
7.6
4
Fold: 4'-phosphopantetheinyl transferaseSuperfamily: 4'-phosphopantetheinyl transferaseFamily: Holo-(acyl carrier protein) synthase ACPS22 d1nkua_
not modelled
7.3
14
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: 3-Methyladenine DNA glycosylase I (Tag)23 c2cg5A_
not modelled
7.1
15
PDB header: transferase/hydrolaseChain: A: PDB Molecule: l-aminoadipate-semialdehyde dehydrogenase-PDBTitle: structure of aminoadipate-semialdehyde dehydrogenase-2 phosphopantetheinyl transferase in complex with cytosolic3 acyl carrier protein and coenzyme a
24 d2fq4a1
not modelled
7.0
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain25 c1u78A_
not modelled
7.0
14
PDB header: dna binding protein/dnaChain: A: PDB Molecule: transposable element tc3 transposase;PDBTitle: structure of the bipartite dna-binding domain of tc32 transposase bound to transposon dna
26 c2osqA_
not modelled
6.8
16
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: nmr structure of rrm-1 of yeast npl3 protein
27 c1bcoA_
not modelled
6.2
13
PDB header: transposaseChain: A: PDB Molecule: bacteriophage mu transposase;PDBTitle: bacteriophage mu transposase core domain
28 d1wf1a_
not modelled
6.2
7
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD29 d1x5ua1
not modelled
6.1
9
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD30 c2xnqA_
not modelled
6.0
20
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein 3;PDBTitle: structural insights into cis element recognition of non-2 polyadenylated rnas by the nab3-rrm
31 c3d1nK_
not modelled
5.9
18
PDB header: transcription regulator/dnaChain: K: PDB Molecule: pou domain, class 6, transcription factor 1;PDBTitle: structure of human brn-5 transcription factor in complex2 with corticotrophin-releasing hormone gene promoter
32 d1ro5a_
not modelled
5.8
16
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: Autoinducer synthetase33 c2jg6A_
not modelled
5.6
7
PDB header: hydrolaseChain: A: PDB Molecule: dna-3-methyladenine glycosidase;PDBTitle: crystal structure of a 3-methyladenine dna glycosylase i2 from staphylococcus aureus
34 d1wg1a_
not modelled
5.6
25
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD35 c3nnhA_
not modelled
5.5
8
PDB header: rna binding protein/rnaChain: A: PDB Molecule: cugbp elav-like family member 1;PDBTitle: crystal structure of the cugbp1 rrm1 with guuguuuuguuu rna
36 c2jvoA_
not modelled
5.3
15
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: segmental isotope labeling of npl3