| 1 |
|
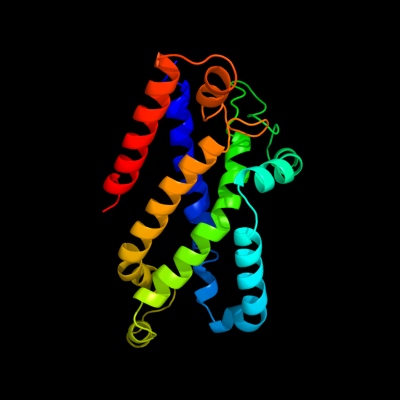
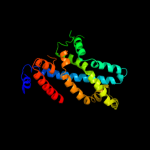
PDB 3dhw chain A domain 1
Region: 93 - 296
Aligned: 194
Modelled: 204
Confidence: 99.9%
Identity: 15%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 2 |
|
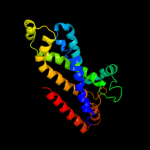
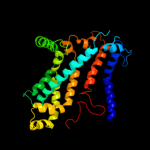
PDB 2onk chain C
Region: 64 - 298
Aligned: 233
Modelled: 235
Confidence: 99.8%
Identity: 12%
PDB header:membrane protein
Chain: C: PDB Molecule:molybdate/tungstate abc transporter, permease
PDBTitle: abc transporter modbc in complex with its binding protein2 moda
Phyre2
| 3 |
|
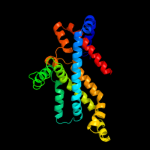
PDB 2onk chain C domain 1
Region: 64 - 298
Aligned: 233
Modelled: 235
Confidence: 99.8%
Identity: 12%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 4 |
|
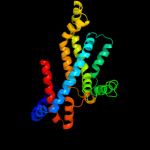
PDB 3d31 chain C domain 1
Region: 92 - 295
Aligned: 202
Modelled: 204
Confidence: 99.8%
Identity: 16%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 5 |
|
PDB 3d31 chain D
Region: 92 - 295
Aligned: 202
Modelled: 204
Confidence: 99.8%
Identity: 16%
PDB header:transport protein
Chain: D: PDB Molecule:sulfate/molybdate abc transporter, permease
PDBTitle: modbc from methanosarcina acetivorans
Phyre2
| 6 |
|
PDB 3fh6 chain F
Region: 1 - 312
Aligned: 284
Modelled: 311
Confidence: 99.7%
Identity: 12%
PDB header:transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
Phyre2
| 7 |
|
PDB 2r6g chain F domain 2
Region: 78 - 294
Aligned: 215
Modelled: 217
Confidence: 99.7%
Identity: 10%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 8 |
|
PDB 2r6g chain F
Region: 72 - 294
Aligned: 221
Modelled: 223
Confidence: 99.7%
Identity: 9%
PDB header:hydrolase/transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: the crystal structure of the e. coli maltose transporter
Phyre2
| 9 |
|
PDB 2r6g chain G domain 1
Region: 6 - 314
Aligned: 270
Modelled: 284
Confidence: 99.4%
Identity: 16%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 10 |
|
PDB 1cii chain A
Region: 77 - 134
Aligned: 55
Modelled: 58
Confidence: 17.1%
Identity: 7%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 11 |
|
PDB 1wz4 chain A
Region: 299 - 307
Aligned: 9
Modelled: 9
Confidence: 13.8%
Identity: 67%
PDB header:gene regulation
Chain: A: PDB Molecule:major surface antigen;
PDBTitle: solution conformation of adr subtype hbv pre-s2 epitope
Phyre2
| 12 |
|
PDB 2cw1 chain A
Region: 198 - 217
Aligned: 18
Modelled: 20
Confidence: 10.3%
Identity: 28%
PDB header:de novo protein
Chain: A: PDB Molecule:sn4m;
PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
Phyre2
| 13 |
|
PDB 2jwa chain A
Region: 100 - 129
Aligned: 30
Modelled: 30
Confidence: 6.8%
Identity: 20%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 14 |
|
PDB 1tdp chain A
Region: 208 - 238
Aligned: 31
Modelled: 31
Confidence: 6.5%
Identity: 19%
Fold: Bromodomain-like
Superfamily: Bacteriocin immunity protein-like
Family: Carnobacteriocin B2 immunity protein
Phyre2
| 15 |
|
PDB 3ci0 chain K domain 2
Region: 294 - 313
Aligned: 20
Modelled: 20
Confidence: 5.5%
Identity: 20%
Fold: SAM domain-like
Superfamily: GspK insert domain-like
Family: GspK insert domain-like
Phyre2
| 16 |
|
PDB 2b9s chain B
Region: 294 - 306
Aligned: 13
Modelled: 13
Confidence: 5.3%
Identity: 31%
PDB header:isomerase/dna
Chain: B: PDB Molecule:dna topoisomerase i-like protein;
PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
Phyre2
| 17 |
|
PDB 2nww chain A domain 1
Region: 3 - 236
Aligned: 189
Modelled: 202
Confidence: 5.3%
Identity: 11%
Fold: Proton glutamate symport protein
Superfamily: Proton glutamate symport protein
Family: Proton glutamate symport protein
Phyre2