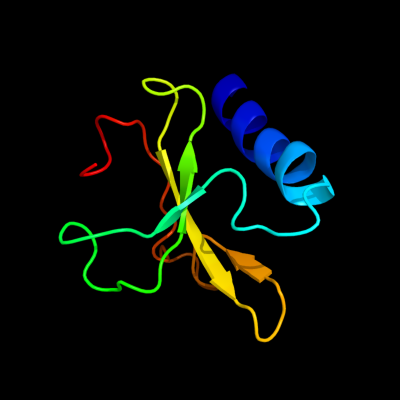
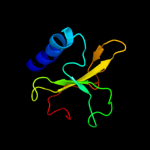
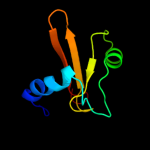
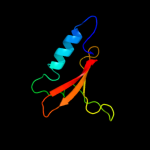
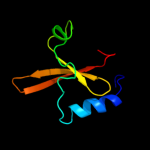
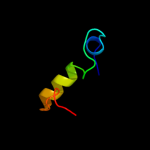
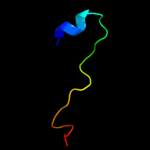
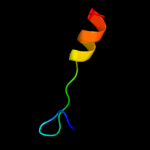
1 c2kheA_
82.1
17
PDB header: hydrolaseChain: A: PDB Molecule: toxin-like protein;PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
2 c2otrA_
63.3
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein hp0892;PDBTitle: solution structure of conserved hypothetical protein hp0892 from2 helicobacter pylori
3 c3g5oC_
62.1
21
PDB header: toxin/antitoxinChain: C: PDB Molecule: uncharacterized protein rv2866;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
4 d1wmia1
52.9
20
Fold: RelE-likeSuperfamily: RelE-likeFamily: RelE-like5 d2a6sa1
35.3
9
Fold: RelE-likeSuperfamily: RelE-likeFamily: YoeB/Txe-like6 d2jeka1
32.2
12
Fold: Rv1873-likeSuperfamily: Rv1873-likeFamily: Rv1873-like7 c2l9vA_
24.4
25
PDB header: rna binding proteinChain: A: PDB Molecule: pre-mrna-processing factor 40 homolog a;PDBTitle: nmr structure of the ff domain l24a mutant's folding transition state
8 d1b79a_
20.1
3
Fold: N-terminal domain of DnaB helicaseSuperfamily: N-terminal domain of DnaB helicaseFamily: N-terminal domain of DnaB helicase9 c2zkr3_
17.8
44
PDB header: ribosomal protein/rnaChain: 3: PDB Molecule: 60s ribosomal protein l39e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
10 d1q46a1
16.9
30
Fold: SAM domain-likeSuperfamily: eIF2alpha middle domain-likeFamily: eIF2alpha middle domain-like11 c3kixy_
16.8
16
PDB header: ribosomeChain: Y: PDB Molecule: PDBTitle: structure of rele nuclease bound to the 70s ribosome2 (postcleavage state; part 3 of 4)
12 c3bpqD_
16.7
10
PDB header: toxinChain: D: PDB Molecule: toxin rele3;PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
13 c3qvaB_
16.6
25
PDB header: hydrolaseChain: B: PDB Molecule: transthyretin-like protein;PDBTitle: structure of klebsiella pneumoniae 5-hydroxyisourate hydrolase
14 d3elna1
16.5
25
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Cysteine dioxygenase type I15 d1jwea_
16.4
3
Fold: N-terminal domain of DnaB helicaseSuperfamily: N-terminal domain of DnaB helicaseFamily: N-terminal domain of DnaB helicase16 d1z8ma1
15.7
17
Fold: RelE-likeSuperfamily: RelE-likeFamily: RelE-like17 d1cmca_
15.4
24
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Met repressor, MetJ (MetR)18 c4a1bB_
15.1
19
PDB header: ribosomeChain: B: PDB Molecule: rpl39;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 3.
19 d1tf5a2
14.6
24
Fold: Helical scaffold and wing domains of SecASuperfamily: Helical scaffold and wing domains of SecAFamily: Helical scaffold and wing domains of SecA20 c3q2bA_
13.4
7
PDB header: actin-binding proteinChain: A: PDB Molecule: cofilin/actin-depolymerizing factor homolog 1;PDBTitle: crystal structure of an actin depolymerizing factor
21 d1rfya_
not modelled
12.9
52
Fold: Long alpha-hairpinSuperfamily: Transcriptional repressor TraMFamily: Transcriptional repressor TraM22 d1nkta2
not modelled
12.8
29
Fold: Helical scaffold and wing domains of SecASuperfamily: Helical scaffold and wing domains of SecAFamily: Helical scaffold and wing domains of SecA23 c3pnxF_
not modelled
12.5
13
PDB header: transferaseChain: F: PDB Molecule: putative sulfurtransferase dsre;PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
24 c3m9hB_
not modelled
12.4
36
PDB header: chaperoneChain: B: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain of the2 mycobacterium tuberculosis proteasomal atpase mpa
25 d1np3a1
not modelled
12.1
21
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Acetohydroxy acid isomeroreductase (ketol-acid reductoisomerase, KARI)26 c1wloA_
not modelled
12.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sufe protein;PDBTitle: solution structure of the hypothetical protein from thermus2 thermophilus hb8
27 d1upgb_
not modelled
11.6
52
Fold: Long alpha-hairpinSuperfamily: Transcriptional repressor TraMFamily: Transcriptional repressor TraM28 c2vzdD_
not modelled
11.6
29
PDB header: cell adhesionChain: D: PDB Molecule: paxillin;PDBTitle: crystal structure of the c-terminal calponin homology2 domain of alpha parvin in complex with paxillin ld1 motif
29 d1m15a2
not modelled
11.5
32
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain30 c3pz8A_
not modelled
11.2
16
PDB header: signaling proteinChain: A: PDB Molecule: segment polarity protein dishevelled homolog dvl-1;PDBTitle: crystal structure of dvl1-dix(y17d) mutant
31 d3ecfa1
not modelled
11.1
46
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ava4193-like32 c3gxvA_
not modelled
11.1
14
PDB header: hydrolase/replicationChain: A: PDB Molecule: replicative dna helicase;PDBTitle: three-dimensional structure of n-terminal domain of dnab2 helicase from helicobacter pylori and its interactions with3 primase
33 c2lhuA_
not modelled
11.1
36
PDB header: structural proteinChain: A: PDB Molecule: mybpc3 protein;PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
34 d2ahob1
not modelled
11.0
41
Fold: SAM domain-likeSuperfamily: eIF2alpha middle domain-likeFamily: eIF2alpha middle domain-like35 d1sqda2
not modelled
11.0
8
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases36 d1u6ra2
not modelled
10.6
25
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain37 c3oeiH_
not modelled
10.6
16
PDB header: toxin, protein bindingChain: H: PDB Molecule: relk (toxin rv3358);PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
38 d1qk1a2
not modelled
10.4
28
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain39 c2rjbD_
not modelled
10.3
8
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein ydcj (sf1787)2 from shigella flexneri which includes domain duf1338.3 northeast structural genomics consortium target sfr276
40 d1crka2
not modelled
10.1
26
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain41 c3l2eB_
not modelled
9.5
32
PDB header: transferaseChain: B: PDB Molecule: glycocyamine kinase beta chain;PDBTitle: glycocyamine kinase, alpha-beta heterodimer from marine worm2 namalycastis sp.
42 d1ni2a3
not modelled
9.4
11
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM43 c3jq3A_
not modelled
9.4
32
PDB header: transferaseChain: A: PDB Molecule: lombricine kinase;PDBTitle: crystal structure of lombricine kinase, complexed with substrate adp
44 d1h4ra3
not modelled
9.3
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM45 d1ef1a3
not modelled
9.3
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM46 c1rl9A_
not modelled
9.2
32
PDB header: transferaseChain: A: PDB Molecule: arginine kinase;PDBTitle: crystal structure of creatine-adp arginine kinase ternary2 complex
47 c1x67A_
not modelled
9.2
16
PDB header: protein bindingChain: A: PDB Molecule: drebrin-like protein;PDBTitle: solution structure of the cofilin homology domain of hip-552 (drebrin-like protein)
48 d1g0wa2
not modelled
9.1
25
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain49 d3cipg1
not modelled
9.0
13
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like50 c3ju6A_
not modelled
8.6
19
PDB header: transferaseChain: A: PDB Molecule: arginine kinase;PDBTitle: crystal structure of dimeric arginine kinase in complex with2 amppnp and arginine
51 d1oo2a_
not modelled
8.6
31
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)52 c2kmaA_
not modelled
8.5
26
PDB header: structural proteinChain: A: PDB Molecule: talin 1;PDBTitle: nmr structure of the f0f1 double domain (residues 1-202) of2 the talin ferm domain
53 c2h1xB_
not modelled
8.4
22
PDB header: hydrolaseChain: B: PDB Molecule: 5-hydroxyisourate hydrolase (formerly known asPDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
54 c4a1aM_
not modelled
8.3
18
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l5;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
55 c2l7qA_
not modelled
8.0
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
56 d1qh4a2
not modelled
7.9
25
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain57 c2r5uD_
not modelled
7.9
10
PDB header: hydrolaseChain: D: PDB Molecule: replicative dna helicase;PDBTitle: crystal structure of the n-terminal domain of dnab helicase from2 mycobacterium tuberculosis
58 c1t0yA_
not modelled
7.5
18
PDB header: chaperoneChain: A: PDB Molecule: tubulin folding cofactor b;PDBTitle: solution structure of a ubiquitin-like domain from tubulin-2 binding cofactor b
59 d1t0ya_
not modelled
7.5
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related60 c3csxA_
not modelled
7.5
40
PDB header: metal binding protein,unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structural characterization of a protein in the duf6832 family- crystal structure of cce_0567 from the3 cyanobacterium cyanothece 51142.
61 d1o66a_
not modelled
7.5
22
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB62 c2fh4C_
not modelled
7.4
19
PDB header: contractile proteinChain: C: PDB Molecule: gelsolin;PDBTitle: c-terminal half of gelsolin soaked in egta at ph 8
63 d1kgia_
not modelled
7.4
35
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)64 d1vrpa2
not modelled
7.3
25
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain65 d1t3ya1
not modelled
7.2
19
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Cofilin-like66 d1qnaa2
not modelled
6.9
13
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain67 d1tdha3
not modelled
6.7
14
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins68 d2bo4a1
not modelled
6.7
45
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: MGS-like69 c3osqA_
not modelled
6.6
36
PDB header: fluorescent protein, transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, green fluorescentPDBTitle: maltose-bound maltose sensor engineered by insertion of circularly2 permuted green fluorescent protein into e. coli maltose binding3 protein at position 175
70 c3pz7A_
not modelled
6.6
18
PDB header: signaling proteinChain: A: PDB Molecule: dixin;PDBTitle: crystal structure of ccd1-dix domain
71 d2djfa1
not modelled
6.5
21
Fold: Streptavidin-likeSuperfamily: Dipeptidyl peptidase I (cathepsin C), exclusion domainFamily: Dipeptidyl peptidase I (cathepsin C), exclusion domain72 c2js5B_
not modelled
6.4
27
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of protein q60c73_metca. northeast structural2 genomics consortium target mcr1
73 c1t0fC_
not modelled
6.2
57
PDB header: dna binding proteinChain: C: PDB Molecule: transposon tn7 transposition protein tnsc;PDBTitle: crystal structure of the tnsa/tnsc(504-555) complex
74 d1udma_
not modelled
6.1
18
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Cofilin-like75 c2kj6A_
not modelled
6.1
18
PDB header: chaperoneChain: A: PDB Molecule: tubulin folding cofactor b;PDBTitle: nmr solution structure of a tubulin folding cofactor b2 obtained from arabidopsis thaliana: northeast structural3 genomics consortium target ar3436a
76 d1cjxa2
not modelled
6.0
31
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases77 d2g3ra1
not modelled
6.0
67
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain78 d1uzca_
not modelled
5.9
27
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain79 d2zpya3
not modelled
5.7
11
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM80 d1d0na4
not modelled
5.7
19
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like81 d1tfpa_
not modelled
5.6
31
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)82 d2fh1a1
not modelled
5.6
15
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like83 d2f7fa1
not modelled
5.6
20
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like84 c3k1lA_
not modelled
5.6
26
PDB header: ligaseChain: A: PDB Molecule: fancl;PDBTitle: crystal structure of fancl
85 c2rooA_
not modelled
5.4
20
PDB header: toxinChain: A: PDB Molecule: neurotoxin magi-4;PDBTitle: solution structure of magi4, a spider toxin from macrothele2 gigas
86 d1f86a_
not modelled
5.4
27
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)87 d1kl9a1
not modelled
5.1
23
Fold: SAM domain-likeSuperfamily: eIF2alpha middle domain-likeFamily: eIF2alpha middle domain-like88 c1i0eD_
not modelled
5.1
25
PDB header: transferaseChain: D: PDB Molecule: creatine kinase,m chain;PDBTitle: crystal structure of creatine kinase from human muscle