1 c1nfoA_
90.9
12
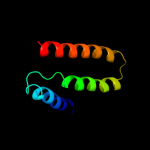
PDB header: lipid transportChain: A: PDB Molecule: apolipoprotein e2;PDBTitle: apolipoprotein e2 (apoe2, d154a mutation)
2 d1gs9a_
86.5
11
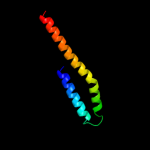
Fold: Four-helical up-and-down bundleSuperfamily: ApolipoproteinFamily: Apolipoprotein3 c3kdpH_
73.7
27
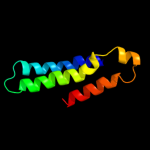
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
4 c3kdpG_
73.7
27
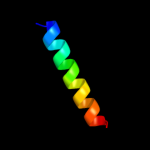
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
5 c2x43S_
65.8
10
PDB header: membrane proteinChain: S: PDB Molecule: sherp;PDBTitle: structural basis of molecular recognition by sherp at membrane2 surfaces
6 c3r2pA_
52.3
8
PDB header: lipid transportChain: A: PDB Molecule: apolipoprotein a-i;PDBTitle: 2.2 angstrom crystal structure of c terminal truncated human2 apolipoprotein a-i reveals the assembly of hdl by dimerization.
7 c2vs0B_
42.6
11
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
8 d1wa8b1
40.8
10
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like9 c2a01B_
39.6
18
PDB header: lipid transportChain: B: PDB Molecule: apolipoprotein a-i;PDBTitle: crystal structure of lipid-free human apolipoprotein a-i
10 c3gvmA_
39.2
6
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
11 d1eq1a_
29.7
14
Fold: Apolipophorin-IIISuperfamily: Apolipophorin-IIIFamily: Apolipophorin-III12 d1wa8a1
25.2
11
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like13 c2jwaA_
21.9
36
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
14 c3onjA_
21.8
11
PDB header: protein transportChain: A: PDB Molecule: t-snare vti1;PDBTitle: crystal structure of yeast vti1p_habc domain
15 c1av1B_
21.8
7
PDB header: lipid transportChain: B: PDB Molecule: apolipoprotein a-i;PDBTitle: crystal structure of human apolipoprotein a-i
16 c1p58F_
21.4
18
PDB header: virusChain: F: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
17 d1f16a_
21.3
21
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death18 c3lw5K_
20.0
44
PDB header: photosynthesisChain: K: PDB Molecule: photosystem i reaction center subunit x psak;PDBTitle: improved model of plant photosystem i
19 c3e0sA_
18.5
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from2 chlorobium tepidum
20 c2l5bA_
16.5
33
PDB header: apoptosisChain: A: PDB Molecule: activator of apoptosis harakiri;PDBTitle: solution structure of the transmembrane domain of bcl-2 member2 harakiri in micelles
21 d1nekd_
not modelled
16.2
15
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)22 d1l2pa_
not modelled
15.5
16
Fold: Single transmembrane helixSuperfamily: F1F0 ATP synthase subunit B, membrane domainFamily: F1F0 ATP synthase subunit B, membrane domain23 d1vcsa1
not modelled
13.9
7
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins24 d1i6la_
not modelled
13.8
4
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain25 c2v8sV_
not modelled
12.3
10
PDB header: protein transportChain: V: PDB Molecule: vesicle transport through interaction withPDBTitle: vti1b habc domain - epsinr enth domain complex
26 c2kncB_
not modelled
12.3
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
27 c2p2uA_
not modelled
12.2
24
PDB header: dna binding proteinChain: A: PDB Molecule: host-nuclease inhibitor protein gam, putative;PDBTitle: crystal structure of putative host-nuclease inhibitor2 protein gam from desulfovibrio vulgaris
28 c3m91B_
not modelled
10.7
17
PDB header: hydrolase regulatorChain: B: PDB Molecule: prokaryotic ubiquitin-like protein pup;PDBTitle: crystal structure of the prokaryotic ubiquitin-like protein (pup)2 complexed with the amino terminal coiled coil of the mycobacterium3 tuberculosis proteasomal atpase mpa
29 d1fioa_
not modelled
10.2
14
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins30 d1ryka_
not modelled
10.1
9
Fold: SAM domain-likeSuperfamily: Hypothetical protein YjbJFamily: Hypothetical protein YjbJ31 c2ww9B_
not modelled
8.9
14
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sss1;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
32 d1s35a1
not modelled
8.8
9
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat33 c2j5dA_
not modelled
8.4
16
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
34 d1eiya1
not modelled
8.3
21
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Phenylalanyl-tRNA synthetase (PheRS)35 d1c99a_
not modelled
7.4
28
Fold: Transmembrane helix hairpinSuperfamily: F1F0 ATP synthase subunit CFamily: F1F0 ATP synthase subunit C36 d1q2ha_
not modelled
7.1
10
Fold: Dimerisation interlockSuperfamily: Phenylalanine zipperFamily: Adapter protein APS, dimerisation domain37 c2kp6A_
not modelled
6.9
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein cv0237 from2 chromobacterium violaceum. northeast structural genomics3 consortium (nesg) target cvt1
38 c1n54A_
not modelled
6.7
9
PDB header: rna binding proteinChain: A: PDB Molecule: 80 kda nuclear cap binding protein;PDBTitle: cap binding complex m7gpppg free
39 c3a0hX_
not modelled
6.7
44
PDB header: electron transportChain: X: PDB Molecule: photosystem ii reaction center protein x;PDBTitle: crystal structure of i-substituted photosystem ii complex
40 c3a0bX_
not modelled
6.7
44
PDB header: electron transportChain: X: PDB Molecule: photosystem ii reaction center protein x;PDBTitle: crystal structure of br-substituted photosystem ii complex
41 c3a0hx_
not modelled
6.7
44
PDB header: electron transportChain: X: PDB Molecule: photosystem ii reaction center protein x;PDBTitle: crystal structure of i-substituted photosystem ii complex
42 c3a0bx_
not modelled
6.7
44
PDB header: electron transportChain: X: PDB Molecule: photosystem ii reaction center protein x;PDBTitle: crystal structure of br-substituted photosystem ii complex
43 c3ipdB_
not modelled
6.5
11
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
44 c3prhB_
not modelled
6.5
9
PDB header: ligaseChain: B: PDB Molecule: tryptophanyl-trna synthetase;PDBTitle: tryptophanyl-trna synthetase val144pro mutant from b. subtilis
45 c2kbvA_
not modelled
6.4
20
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
46 d1lvfa_
not modelled
6.3
8
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins47 c3sz3A_
not modelled
6.1
12
PDB header: ligaseChain: A: PDB Molecule: tryptophanyl-trna synthetase;PDBTitle: crystal structure of tryptophanyl-trna synthetase from vibrio cholerae2 with an endogenous tryptophan
48 c2ka1B_
not modelled
6.1
16
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
49 c2ka2A_
not modelled
6.1
16
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
50 c1s5lx_
not modelled
6.0
44
PDB header: photosynthesisChain: X: PDB Molecule: photosystem ii psbx protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
51 d1dqna_
not modelled
5.8
16
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)52 c2ka1A_
not modelled
5.7
13
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
53 c2ka2B_
not modelled
5.7
13
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
54 d1rhzb_
not modelled
5.7
21
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit55 c1q90L_
not modelled
5.6
33
PDB header: photosynthesisChain: L: PDB Molecule: cytochrome b6f complex subunit petl;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
56 d1q90l_
not modelled
5.6
33
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex57 c3hd7A_
not modelled
5.4
15
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
58 d1t72a_
not modelled
5.4
13
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like59 c3py7A_
not modelled
5.4
18
PDB header: viral proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,paxillin ld1,protein e6PDBTitle: crystal structure of full-length bovine papillomavirus oncoprotein e62 in complex with ld1 motif of paxillin at 2.3a resolution