| 1 |
|
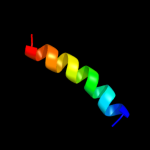
PDB 1ijd chain B
Region: 7 - 20
Aligned: 14
Modelled: 14
Confidence: 19.6%
Identity: 64%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 2 |
|
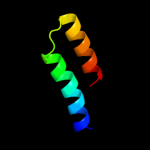
PDB 1jb0 chain M
Region: 9 - 20
Aligned: 12
Modelled: 12
Confidence: 17.6%
Identity: 67%
PDB header:photosynthesis
Chain: M: PDB Molecule:photosystem 1 reaction centre subunit xii;
PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
Phyre2
| 3 |
|
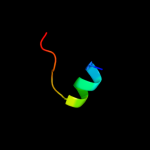
PDB 1jb0 chain M
Region: 9 - 20
Aligned: 12
Modelled: 12
Confidence: 17.6%
Identity: 67%
Fold: Single transmembrane helix
Superfamily: Subunit XII of photosystem I reaction centre, PsaM
Family: Subunit XII of photosystem I reaction centre, PsaM
Phyre2
| 4 |
|
PDB 2iub chain A domain 2
Region: 4 - 36
Aligned: 33
Modelled: 33
Confidence: 15.3%
Identity: 24%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 5 |
|
PDB 1xme chain B domain 2
Region: 2 - 15
Aligned: 14
Modelled: 14
Confidence: 12.2%
Identity: 50%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 6 |
|
PDB 2ari chain A
Region: 15 - 31
Aligned: 17
Modelled: 17
Confidence: 9.2%
Identity: 59%
PDB header:viral protein
Chain: A: PDB Molecule:envelope polyprotein gp160;
PDBTitle: solution structure of micelle-bound fusion domain of hiv-12 gp41
Phyre2
| 7 |
|
PDB 2klu chain A
Region: 16 - 36
Aligned: 21
Modelled: 21
Confidence: 8.2%
Identity: 29%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 8 |
|
PDB 1rh1 chain A domain 2
Region: 3 - 39
Aligned: 37
Modelled: 37
Confidence: 7.3%
Identity: 24%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 9 |
|
PDB 1erf chain A
Region: 15 - 31
Aligned: 17
Modelled: 17
Confidence: 6.1%
Identity: 53%
PDB header:viral protein
Chain: A: PDB Molecule:transmembrane glycoprotein;
PDBTitle: conformational mapping of the n-terminal fusion peptide of2 hiv-1 gp41 using 13c-enhanced fourier transform infrared3 spectroscopy (ftir)
Phyre2
| 10 |
|
PDB 2j7a chain C
Region: 8 - 29
Aligned: 22
Modelled: 22
Confidence: 6.0%
Identity: 5%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome c quinol dehydrogenase nrfh;
PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
Phyre2
| 11 |
|
PDB 3m7b chain A
Region: 15 - 48
Aligned: 34
Modelled: 34
Confidence: 5.8%
Identity: 21%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tellurite resistance protein teha homolog;
PDBTitle: crystal structure of plant slac1 homolog teha
Phyre2
| 12 |
|
PDB 1cii chain A
Region: 16 - 49
Aligned: 30
Modelled: 34
Confidence: 5.8%
Identity: 27%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 13 |
|
PDB 2bbj chain B
Region: 4 - 38
Aligned: 35
Modelled: 35
Confidence: 5.6%
Identity: 26%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2