| 1 |
|
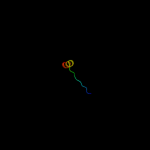
PDB 1n7v chain A
Region: 121 - 141
Aligned: 21
Modelled: 21
Confidence: 17.9%
Identity: 24%
Fold: Adsorption protein p2
Superfamily: Adsorption protein p2
Family: Adsorption protein p2
Phyre2
| 2 |
|
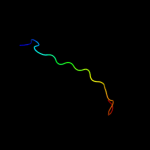
PDB 3kys chain C
Region: 125 - 144
Aligned: 20
Modelled: 20
Confidence: 10.1%
Identity: 35%
PDB header:transcription/protein binding
Chain: C: PDB Molecule:transcriptional enhancer factor tef-1;
PDBTitle: crystal structure of human yap and tead complex
Phyre2
| 3 |
|
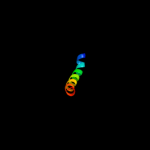
PDB 3l15 chain A
Region: 125 - 144
Aligned: 20
Modelled: 20
Confidence: 9.2%
Identity: 25%
PDB header:transcription
Chain: A: PDB Molecule:transcriptional enhancer factor tef-4;
PDBTitle: human tead2 transcriptional factor
Phyre2
| 4 |
|
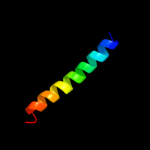
PDB 2bbg chain A
Region: 116 - 134
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 21%
Fold: Amb V allergen
Superfamily: Amb V allergen
Family: Amb V allergen
Phyre2
| 5 |
|
PDB 2cpb chain A
Region: 18 - 43
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 8%
PDB header:viral protein
Chain: A: PDB Molecule:m13 major coat protein;
PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
Phyre2
| 6 |
|
PDB 2knc chain A
Region: 88 - 116
Aligned: 29
Modelled: 29
Confidence: 6.8%
Identity: 17%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 7 |
|
PDB 3lu2 chain B
Region: 49 - 59
Aligned: 11
Modelled: 11
Confidence: 5.7%
Identity: 36%
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2462 protein;
PDBTitle: structure of lmo2462, a listeria monocytogenes amidohydrolase family2 putative dipeptidase
Phyre2
| 8 |
|
PDB 1itu chain A
Region: 49 - 59
Aligned: 11
Modelled: 11
Confidence: 5.2%
Identity: 27%
Fold: TIM beta/alpha-barrel
Superfamily: Metallo-dependent hydrolases
Family: Renal dipeptidase
Phyre2