| 1 |
|
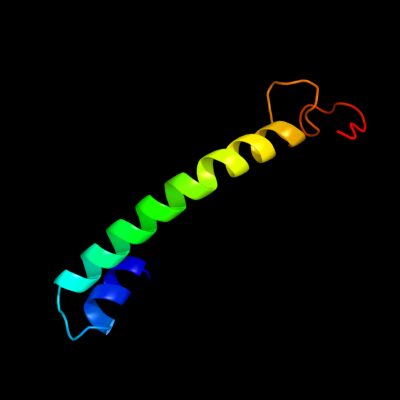
PDB 3qnq chain D
Region: 208 - 266
Aligned: 54
Modelled: 59
Confidence: 65.9%
Identity: 11%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 2 |
|
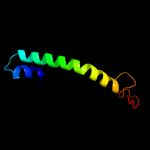
PDB 3iz5 chain Q
Region: 236 - 266
Aligned: 31
Modelled: 31
Confidence: 40.0%
Identity: 29%
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 3 |
|
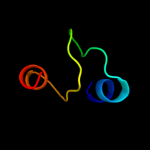
PDB 2l34 chain B
Region: 30 - 64
Aligned: 33
Modelled: 34
Confidence: 37.3%
Identity: 30%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 4 |
|
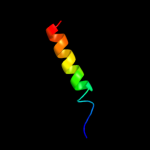
PDB 2l35 chain B
Region: 30 - 52
Aligned: 23
Modelled: 23
Confidence: 34.6%
Identity: 30%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 5 |
|
PDB 2l34 chain A
Region: 36 - 52
Aligned: 17
Modelled: 17
Confidence: 27.1%
Identity: 41%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 6 |
|
PDB 2l35 chain A
Region: 36 - 65
Aligned: 28
Modelled: 30
Confidence: 19.2%
Identity: 36%
PDB header:protein binding
Chain: A: PDB Molecule:dap12-nkg2c_tm;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 7 |
|
PDB 1kqf chain B domain 2
Region: 223 - 264
Aligned: 32
Modelled: 42
Confidence: 14.9%
Identity: 28%
Fold: Single transmembrane helix
Superfamily: Iron-sulfur subunit of formate dehydrogenase N, transmembrane anchor
Family: Iron-sulfur subunit of formate dehydrogenase N, transmembrane anchor
Phyre2
| 8 |
|
PDB 1cii chain A
Region: 69 - 120
Aligned: 51
Modelled: 52
Confidence: 14.5%
Identity: 14%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 9 |
|
PDB 2knc chain A
Region: 224 - 264
Aligned: 35
Modelled: 41
Confidence: 7.4%
Identity: 20%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 10 |
|
PDB 1r7m chain A domain 1
Region: 32 - 45
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 29%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 11 |
|
PDB 1r7m chain A
Region: 32 - 45
Aligned: 14
Modelled: 14
Confidence: 5.7%
Identity: 29%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:intron-encoded endonuclease i-scei;
PDBTitle: the homing endonuclease i-scei bound to its dna recognition2 region
Phyre2