| 1 |
|
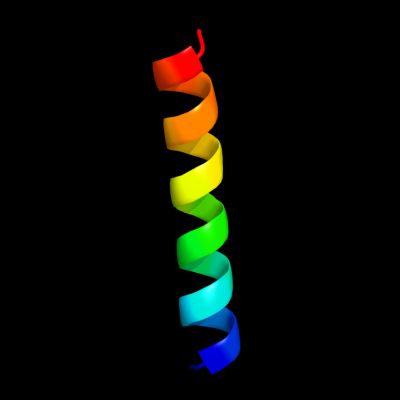
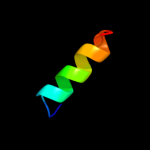
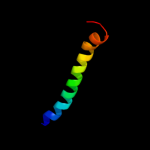
PDB 2jok chain A domain 1
Region: 14 - 35
Aligned: 22
Modelled: 22
Confidence: 10.7%
Identity: 32%
Fold: SopE-like GEF domain
Superfamily: SopE-like GEF domain
Family: SopE-like GEF domain
Phyre2
| 2 |
|
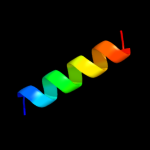
PDB 2kel chain B
Region: 22 - 29
Aligned: 8
Modelled: 8
Confidence: 9.7%
Identity: 50%
PDB header:transcription repressor
Chain: B: PDB Molecule:uncharacterized protein 56b;
PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
Phyre2
| 3 |
|
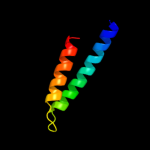
PDB 1sg7 chain A
Region: 51 - 66
Aligned: 16
Modelled: 16
Confidence: 8.4%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cation transport regulator chab;
PDBTitle: nmr solution structure of the putative cation transport2 regulator chab
Phyre2
| 4 |
|
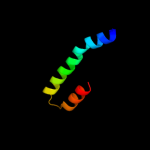
PDB 1sg7 chain A domain 1
Region: 51 - 66
Aligned: 16
Modelled: 16
Confidence: 8.4%
Identity: 19%
Fold: ChaB-like
Superfamily: ChaB-like
Family: ChaB-like
Phyre2
| 5 |
|
PDB 3swf chain A
Region: 8 - 33
Aligned: 26
Modelled: 26
Confidence: 7.5%
Identity: 23%
PDB header:transport protein
Chain: A: PDB Molecule:cgmp-gated cation channel alpha-1;
PDBTitle: cnga1 621-690 containing clz domain
Phyre2
| 6 |
|
PDB 1wnk chain A
Region: 42 - 56
Aligned: 15
Modelled: 15
Confidence: 7.3%
Identity: 53%
PDB header:transcription
Chain: A: PDB Molecule:fibroin-modulator-binding-protein-1;
PDBTitle: nmr structure of fmbp-1 tandem repeat 3 in 30%(v/v) tfe2 solution
Phyre2
| 7 |
|
PDB 1s35 chain A domain 2
Region: 6 - 48
Aligned: 43
Modelled: 43
Confidence: 7.0%
Identity: 9%
Fold: Spectrin repeat-like
Superfamily: Spectrin repeat
Family: Spectrin repeat
Phyre2
| 8 |
|
PDB 1ka8 chain A
Region: 47 - 63
Aligned: 17
Modelled: 17
Confidence: 6.6%
Identity: 41%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: P4 origin-binding domain-like
Phyre2
| 9 |
|
PDB 3t38 chain B
Region: 40 - 56
Aligned: 17
Modelled: 17
Confidence: 6.4%
Identity: 24%
PDB header:oxidoreductase
Chain: B: PDB Molecule:arsenate reductase;
PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
Phyre2
| 10 |
|
PDB 1u5p chain A domain 1
Region: 6 - 55
Aligned: 50
Modelled: 50
Confidence: 6.3%
Identity: 16%
Fold: Spectrin repeat-like
Superfamily: Spectrin repeat
Family: Spectrin repeat
Phyre2
| 11 |
|
PDB 1u5p chain A domain 2
Region: 6 - 48
Aligned: 43
Modelled: 43
Confidence: 6.2%
Identity: 19%
Fold: Spectrin repeat-like
Superfamily: Spectrin repeat
Family: Spectrin repeat
Phyre2
| 12 |
|
PDB 2spc chain A
Region: 6 - 56
Aligned: 51
Modelled: 51
Confidence: 6.1%
Identity: 8%
Fold: Spectrin repeat-like
Superfamily: Spectrin repeat
Family: Spectrin repeat
Phyre2
| 13 |
|
PDB 1hci chain A domain 4
Region: 6 - 51
Aligned: 46
Modelled: 46
Confidence: 5.9%
Identity: 11%
Fold: Spectrin repeat-like
Superfamily: Spectrin repeat
Family: Spectrin repeat
Phyre2
| 14 |
|
PDB 1wlx chain A
Region: 6 - 39
Aligned: 34
Modelled: 34
Confidence: 5.3%
Identity: 6%
PDB header:protein binding
Chain: A: PDB Molecule:alpha-actinin 4;
PDBTitle: solution structure of the third spectrin repeat of alpha-2 actinin-4
Phyre2