| 1 |
|
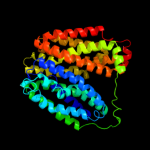
PDB 3qe7 chain A
Region: 17 - 443
Aligned: 397
Modelled: 427
Confidence: 100.0%
Identity: 28%
PDB header:transport protein
Chain: A: PDB Molecule:uracil permease;
PDBTitle: crystal structure of uracil transporter--uraa
Phyre2
| 2 |
|
PDB 1pw4 chain A
Region: 8 - 458
Aligned: 424
Modelled: 430
Confidence: 96.1%
Identity: 9%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
PDB 3lpz chain A
Region: 299 - 315
Aligned: 17
Modelled: 17
Confidence: 14.2%
Identity: 18%
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
Phyre2
| 4 |
|
PDB 1pv7 chain A
Region: 28 - 456
Aligned: 406
Modelled: 412
Confidence: 11.1%
Identity: 8%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 5 |
|
PDB 2rng chain A
Region: 465 - 481
Aligned: 17
Modelled: 17
Confidence: 10.1%
Identity: 29%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:big defensin;
PDBTitle: solution structure of big defensin
Phyre2
| 6 |
|
PDB 3org chain B
Region: 288 - 444
Aligned: 157
Modelled: 157
Confidence: 7.9%
Identity: 15%
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
Phyre2
| 7 |
|
PDB 2xut chain C
Region: 27 - 210
Aligned: 179
Modelled: 184
Confidence: 7.6%
Identity: 9%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 8 |
|
PDB 3dla chain D
Region: 413 - 480
Aligned: 68
Modelled: 68
Confidence: 7.5%
Identity: 10%
PDB header:ligase
Chain: D: PDB Molecule:glutamine-dependent nad(+) synthetase;
PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
Phyre2
| 9 |
|
PDB 1dxz chain A
Region: 23 - 46
Aligned: 24
Modelled: 24
Confidence: 7.5%
Identity: 13%
PDB header:transmembrane protein
Chain: A: PDB Molecule:acetylcholine receptor protein, alpha chain;
PDBTitle: m2 transmembrane segment of alpha-subunit of nicotinic2 acetylcholine receptor from torpedo californica, nmr, 203 structures
Phyre2
| 10 |
|
PDB 1oed chain B
Region: 389 - 480
Aligned: 91
Modelled: 92
Confidence: 7.2%
Identity: 14%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 11 |
|
PDB 2la2 chain A
Region: 468 - 481
Aligned: 14
Modelled: 14
Confidence: 6.9%
Identity: 21%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:cecropin;
PDBTitle: solution structure of papiliocin isolated from the swallowtail2 butterfly, papilio xuthus
Phyre2
| 12 |
|
PDB 2knc chain B
Region: 461 - 482
Aligned: 22
Modelled: 22
Confidence: 6.2%
Identity: 9%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 13 |
|
PDB 2gfp chain A
Region: 30 - 437
Aligned: 367
Modelled: 377
Confidence: 6.1%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 14 |
|
PDB 2oar chain A
Region: 339 - 474
Aligned: 112
Modelled: 115
Confidence: 6.1%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 15 |
|
PDB 3hd6 chain A
Region: 300 - 457
Aligned: 157
Modelled: 158
Confidence: 5.9%
Identity: 10%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 16 |
|
PDB 3dhw chain A domain 1
Region: 350 - 474
Aligned: 125
Modelled: 125
Confidence: 5.8%
Identity: 7%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 17 |
|
PDB 1u0s chain A
Region: 458 - 477
Aligned: 20
Modelled: 20
Confidence: 5.5%
Identity: 20%
Fold: Ferredoxin-like
Superfamily: CheY-binding domain of CheA
Family: CheY-binding domain of CheA
Phyre2
| 18 |
|
PDB 2nny chain A
Region: 446 - 482
Aligned: 37
Modelled: 37
Confidence: 5.4%
Identity: 5%
PDB header:transcription/dna
Chain: A: PDB Molecule:c-ets-1 protein;
PDBTitle: crystal structure of the ets1 dimer dna complex.
Phyre2
| 19 |
|
PDB 1gvj chain A
Region: 446 - 482
Aligned: 37
Modelled: 37
Confidence: 5.2%
Identity: 5%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: ets domain
Phyre2