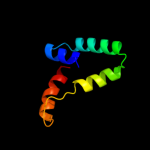
1 c2hh7A_
99.9
26
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein csor;PDBTitle: crystal structure of cu(i) bound csor from mycobacterium tuberculosis.
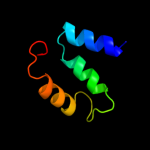
2 c3aaiB_
99.9
22
PDB header: transcriptionChain: B: PDB Molecule: copper homeostasis operon regulatory protein;PDBTitle: x-ray crystal structure of csor from thermus thermophilus hb8
3 c1paqA_
41.5
14
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b epsilonPDBTitle: crystal structure of the catalytic fragment of eukaryotic2 initiation factor 2b epsilon
4 d1paqa_
41.5
14
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: MIF4G domain-like5 c2p5tA_
32.3
44
PDB header: transcription regulatorChain: A: PDB Molecule: putative transcriptional regulator peza;PDBTitle: molecular and structural characterization of the pezat chromosomal2 toxin-antitoxin system of the human pathogen streptococcus pneumoniae
6 c3juiA_
28.5
19
PDB header: translationChain: A: PDB Molecule: translation initiation factor eif-2b subunit epsilon;PDBTitle: crystal structure of the c-terminal domain of human translation2 initiation factor eif2b epsilon subunit
7 d1obba2
22.7
20
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase8 d1ivsa1
19.9
13
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Valyl-tRNA synthetase (ValRS) C-terminal domain9 c3nepX_
19.4
18
PDB header: oxidoreductaseChain: X: PDB Molecule: malate dehydrogenase;PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
10 c3frwF_
19.4
13
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative trp repressor protein;PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
11 c2nv2U_
19.1
18
PDB header: lyase/transferaseChain: U: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
12 c3zs9D_
17.6
44
PDB header: hydrolase/transport proteinChain: D: PDB Molecule: golgi to er traffic protein 2;PDBTitle: s. cerevisiae get3-adp-alf4- complex with a cytosolic get2 fragment
13 d1up7a2
17.6
10
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: AglA-like glucosidase14 c2qycA_
16.5
23
PDB header: unknown functionChain: A: PDB Molecule: ferredoxin-like protein;PDBTitle: crystal structure of a dimeric ferredoxin-like protein (bb1511) from2 bordetella bronchiseptica rb50 at 1.90 a resolution
15 d1ev0a_
14.1
25
Fold: Cell division protein MinE topological specificity domainSuperfamily: Cell division protein MinE topological specificity domainFamily: Cell division protein MinE topological specificity domain16 c3cveC_
11.8
25
PDB header: signaling proteinChain: C: PDB Molecule: homer protein homolog 1;PDBTitle: crystal structure of the carboxy terminus of homer1
17 c3korD_
11.1
16
PDB header: transcriptionChain: D: PDB Molecule: possible trp repressor;PDBTitle: crystal structure of a putative trp repressor from staphylococcus2 aureus
18 c2gtlO_
10.2
19
PDB header: oxygen storage/transportChain: O: PDB Molecule: extracellular hemoglobin linker l3 subunit;PDBTitle: lumbricus erythrocruorin at 3.5a resolution
19 c4a1qB_
9.8
16
PDB header: viral proteinChain: B: PDB Molecule: orf e73;PDBTitle: solution structure of e73 protein from sulfolobus spindle-2 shaped virus ragged hills, a hyperthermophilic3 crenarchaeal virus from yellowstone national park
20 c3femB_
9.0
24
PDB header: biosynthetic protein, transferaseChain: B: PDB Molecule: pyridoxine biosynthesis protein snz1;PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
21 c3ku7B_
not modelled
8.3
25
PDB header: cell cycleChain: B: PDB Molecule: cell division topological specificity factor;PDBTitle: crystal structure of helicobacter pylori mine, a cell division2 topological specificity factor
22 c1obbB_
not modelled
8.0
20
PDB header: hydrolaseChain: B: PDB Molecule: alpha-glucosidase;PDBTitle: alpha-glucosidase a, agla, from thermotoga maritima in2 complex with maltose and nad+
23 d2hyec2
not modelled
7.8
15
Fold: alpha-alpha superhelixSuperfamily: Cullin repeat-likeFamily: Cullin repeat24 d1tr0a_
not modelled
7.8
13
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein25 d1q4ra_
not modelled
7.8
14
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein26 d1sxjd1
not modelled
7.6
5
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: DNA polymerase III clamp loader subunits, C-terminal domain27 d1qnaa2
not modelled
7.2
7
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain28 c1up6F_
not modelled
7.1
10
PDB header: hydrolaseChain: F: PDB Molecule: 6-phospho-beta-glucosidase;PDBTitle: structure of the 6-phospho-beta glucosidase from thermotoga2 maritima at 2.55 angstrom resolution in the tetragonal form3 with manganese, nad+ and glucose-6-phosphate
29 d1cdwa2
not modelled
6.3
7
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain30 d1c2ya_
not modelled
6.2
14
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase31 c2ovcA_
not modelled
6.2
25
PDB header: transport proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily kqt member 4;PDBTitle: crystal structure of a coiled-coil tetramerization domain from kv7.42 channels
32 d1josa_
not modelled
5.8
5
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA33 d1kkga_
not modelled
5.6
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA34 c2xa0C_
not modelled
5.5
35
PDB header: apoptosisChain: C: PDB Molecule: apoptosis regulator bax;PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
35 c3bdeA_
not modelled
5.2
25
PDB header: unknown functionChain: A: PDB Molecule: mll5499 protein;PDBTitle: crystal structure of a dabb family protein with a ferredoxin-like fold2 (mll5499) from mesorhizobium loti maff303099 at 1.79 a resolution
36 c3fajA_
not modelled
5.2
5
PDB header: structural proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of the structural protein p131 of the archaeal virus2 acidianus two-tailed virus (atv)
37 c3hugJ_
not modelled
5.1
23
PDB header: transcription/membrane proteinChain: J: PDB Molecule: probable conserved membrane protein;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
38 c2kxoA_
not modelled
5.0
17
PDB header: cell cycleChain: A: PDB Molecule: cell division topological specificity factor;PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae