| 1 |
|
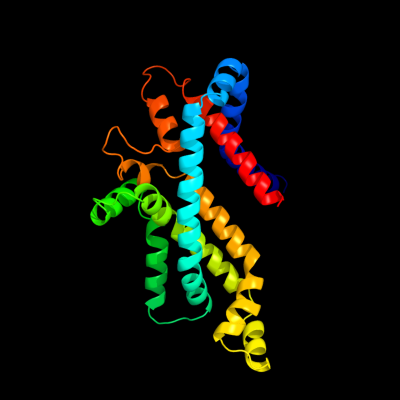
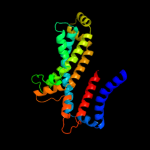
PDB 2onk chain C
Region: 26 - 285
Aligned: 236
Modelled: 242
Confidence: 99.9%
Identity: 17%
PDB header:membrane protein
Chain: C: PDB Molecule:molybdate/tungstate abc transporter, permease
PDBTitle: abc transporter modbc in complex with its binding protein2 moda
Phyre2
| 2 |
|
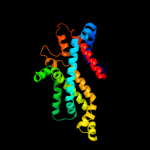
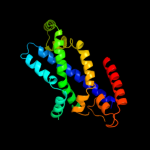
PDB 2onk chain C domain 1
Region: 26 - 285
Aligned: 236
Modelled: 242
Confidence: 99.9%
Identity: 17%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 3 |
|
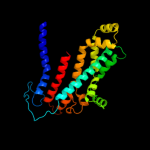
PDB 3fh6 chain F
Region: 21 - 287
Aligned: 267
Modelled: 267
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
Phyre2
| 4 |
|
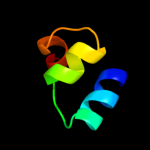
PDB 3dhw chain A domain 1
Region: 89 - 283
Aligned: 187
Modelled: 195
Confidence: 99.9%
Identity: 17%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 5 |
|
PDB 2r6g chain F domain 2
Region: 79 - 281
Aligned: 203
Modelled: 203
Confidence: 99.9%
Identity: 14%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 6 |
|
PDB 3d31 chain C domain 1
Region: 32 - 283
Aligned: 229
Modelled: 235
Confidence: 99.9%
Identity: 14%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 7 |
|
PDB 3d31 chain D
Region: 32 - 283
Aligned: 229
Modelled: 235
Confidence: 99.9%
Identity: 14%
PDB header:transport protein
Chain: D: PDB Molecule:sulfate/molybdate abc transporter, permease
PDBTitle: modbc from methanosarcina acetivorans
Phyre2
| 8 |
|
PDB 2r6g chain F
Region: 88 - 281
Aligned: 194
Modelled: 194
Confidence: 99.9%
Identity: 14%
PDB header:hydrolase/transport protein
Chain: F: PDB Molecule:maltose transport system permease protein malf;
PDBTitle: the crystal structure of the e. coli maltose transporter
Phyre2
| 9 |
|
PDB 2r6g chain G domain 1
Region: 28 - 283
Aligned: 254
Modelled: 256
Confidence: 99.9%
Identity: 15%
Fold: MetI-like
Superfamily: MetI-like
Family: MetI-like
Phyre2
| 10 |
|
PDB 2jwa chain A
Region: 94 - 127
Aligned: 34
Modelled: 34
Confidence: 29.1%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 11 |
|
PDB 1umq chain A
Region: 173 - 204
Aligned: 32
Modelled: 32
Confidence: 9.5%
Identity: 9%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 12 |
|
PDB 1umq chain A
Region: 173 - 204
Aligned: 32
Modelled: 32
Confidence: 9.5%
Identity: 9%
PDB header:dna-binding protein
Chain: A: PDB Molecule:photosynthetic apparatus regulatory protein;
PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
Phyre2
| 13 |
|
PDB 2cw1 chain A
Region: 176 - 197
Aligned: 20
Modelled: 22
Confidence: 7.8%
Identity: 25%
PDB header:de novo protein
Chain: A: PDB Molecule:sn4m;
PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
Phyre2
| 14 |
|
PDB 1wz4 chain A
Region: 289 - 295
Aligned: 7
Modelled: 7
Confidence: 7.3%
Identity: 57%
PDB header:gene regulation
Chain: A: PDB Molecule:major surface antigen;
PDBTitle: solution conformation of adr subtype hbv pre-s2 epitope
Phyre2
| 15 |
|
PDB 1ntc chain A
Region: 173 - 205
Aligned: 33
Modelled: 33
Confidence: 7.1%
Identity: 24%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 16 |
|
PDB 1eto chain B
Region: 173 - 205
Aligned: 33
Modelled: 33
Confidence: 7.1%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 17 |
|
PDB 1fip chain A
Region: 173 - 205
Aligned: 33
Modelled: 33
Confidence: 7.0%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 18 |
|
PDB 1g2h chain A
Region: 189 - 205
Aligned: 17
Modelled: 17
Confidence: 5.7%
Identity: 24%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2
| 19 |
|
PDB 1etx chain A
Region: 173 - 205
Aligned: 33
Modelled: 33
Confidence: 5.3%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: FIS-like
Phyre2