| 1 |
|
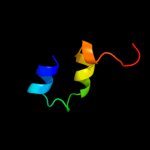
PDB 2l42 chain A
Region: 76 - 105
Aligned: 24
Modelled: 30
Confidence: 12.7%
Identity: 33%
PDB header:protein binding
Chain: A: PDB Molecule:dna-binding protein rap1;
PDBTitle: the solution structure of rap1 brct domain from saccharomyces2 cerevisiae
Phyre2
| 2 |
|
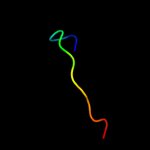
PDB 2bbk chain L
Region: 92 - 104
Aligned: 13
Modelled: 13
Confidence: 9.1%
Identity: 23%
Fold: Methylamine dehydrogenase, L chain
Superfamily: Methylamine dehydrogenase, L chain
Family: Methylamine dehydrogenase, L chain
Phyre2
| 3 |
|
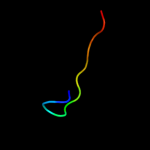
PDB 3c75 chain L
Region: 92 - 104
Aligned: 13
Modelled: 13
Confidence: 8.9%
Identity: 23%
PDB header:oxidoreductase
Chain: L: PDB Molecule:methylamine dehydrogenase light chain;
PDBTitle: paracoccus versutus methylamine dehydrogenase in complex2 with amicyanin
Phyre2
| 4 |
|
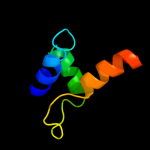
PDB 2iur chain D
Region: 92 - 104
Aligned: 13
Modelled: 13
Confidence: 8.8%
Identity: 23%
PDB header:oxidoreductase
Chain: D: PDB Molecule:aromatic amine dehydrogenase beta subunit;
PDBTitle: crystal structure of n-quinol form of aromatic amine2 dehydrogenase (aadh) from alcaligenes faecalis, form a3 cocrystal
Phyre2
| 5 |
|
PDB 2di0 chain A domain 1
Region: 36 - 92
Aligned: 45
Modelled: 57
Confidence: 8.6%
Identity: 22%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: CUE domain
Phyre2
| 6 |
|
PDB 1fi8 chain F
Region: 94 - 106
Aligned: 13
Modelled: 13
Confidence: 6.7%
Identity: 38%
PDB header:hydrolase/hydrolase inhibitor
Chain: F: PDB Molecule:ecotin;
PDBTitle: rat granzyme b [n66q] complexed to ecotin [81-84 iepd]
Phyre2
| 7 |
|
PDB 2xvc chain A
Region: 118 - 137
Aligned: 20
Modelled: 20
Confidence: 5.2%
Identity: 20%
PDB header:cell cycle
Chain: A: PDB Molecule:escrt-iii;
PDBTitle: molecular and structural basis of escrt-iii recruitment to2 membranes during archaeal cell division
Phyre2