| 1 |
|
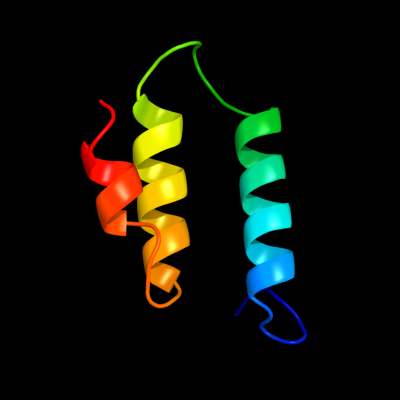
PDB 1cii chain A
Region: 69 - 120
Aligned: 51
Modelled: 52
Confidence: 40.2%
Identity: 16%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 2 |
|
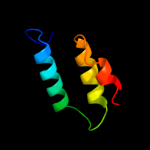
PDB 2l35 chain B
Region: 30 - 52
Aligned: 23
Modelled: 23
Confidence: 37.0%
Identity: 39%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 3 |
|
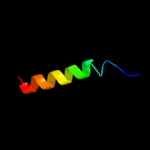
PDB 2l34 chain B
Region: 30 - 52
Aligned: 23
Modelled: 23
Confidence: 30.9%
Identity: 39%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 4 |
|
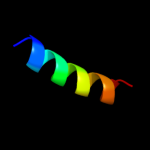
PDB 2l34 chain A
Region: 36 - 52
Aligned: 17
Modelled: 17
Confidence: 28.3%
Identity: 47%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 5 |
|
PDB 2l35 chain A
Region: 36 - 52
Aligned: 17
Modelled: 17
Confidence: 13.0%
Identity: 47%
PDB header:protein binding
Chain: A: PDB Molecule:dap12-nkg2c_tm;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 6 |
|
PDB 3a0b chain X
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 8.3%
Identity: 30%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 7 |
|
PDB 3a0h chain X
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 8.3%
Identity: 30%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 8 |
|
PDB 3a0b chain X
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 8.3%
Identity: 30%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 9 |
|
PDB 3a0h chain X
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 8.3%
Identity: 30%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 10 |
|
PDB 1s5l chain X
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 7.1%
Identity: 30%
PDB header:photosynthesis
Chain: X: PDB Molecule:photosystem ii psbx protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
Phyre2
| 11 |
|
PDB 3fse chain B
Region: 43 - 50
Aligned: 8
Modelled: 8
Confidence: 6.1%
Identity: 75%
PDB header:hydrolase
Chain: B: PDB Molecule:two-domain protein containing dj-1/thij/pfpi-like and
PDBTitle: crystal structure of a two-domain protein containing dj-1/thij/pfpi-2 like and ferritin-like domains (ava_4496) from anabaena variabilis3 atcc 29413 at 1.90 a resolution
Phyre2
| 12 |
|
PDB 1r7m chain A domain 1
Region: 32 - 45
Aligned: 14
Modelled: 14
Confidence: 5.9%
Identity: 36%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2