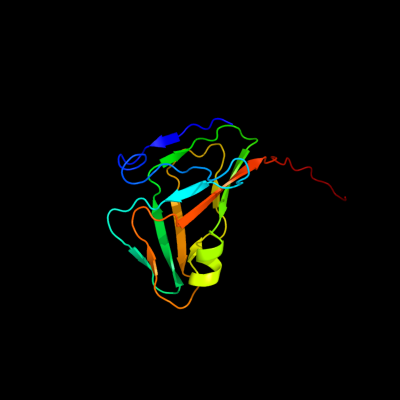
1 c3ehwA_
100.0
32
PDB header: hydrolaseChain: A: PDB Molecule: dutp pyrophosphatase;PDBTitle: human dutpase in complex with alpha,beta-imido-dutp and mg2+:2 visualization of the full-length c-termini in all monomers and3 suggestion for an additional metal ion binding site
2 d1sixa_
100.0
32
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like3 d1rnja_
100.0
99
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like4 d1euwa_
100.0
100
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like5 c3tqzA_
100.0
61
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut)2 from coxiella burnetii
6 d3ehwa1
100.0
32
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like7 c3f4fB_
100.0
29
PDB header: hydrolaseChain: B: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of dut1p, a dutpase from saccharomyces cerevisiae
8 d1f7ra_
100.0
26
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like9 c3mbqC_
100.0
44
PDB header: hydrolaseChain: C: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5-triphosphate nucleotidohydrolase2 from brucella melitensis, orthorhombic crystal form
10 d1sjna_
100.0
34
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like11 c3c3iA_
100.0
31
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine triphosphatase;PDBTitle: evolution of chlorella virus dutpase
12 c3ca9A_
100.0
32
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine triphosphatase;PDBTitle: evolution of chlorella virus dutpase
13 c3lqwA_
100.0
29
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5-triphosphate2 nucleotidohydrolase from entamoeba histolytica
14 d1vyqa1
100.0
26
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like15 d1q5uz_
100.0
29
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like16 c2p9oB_
100.0
31
PDB header: hydrolaseChain: B: PDB Molecule: dutp pyrophosphatase-like protein;PDBTitle: structure of dutpase from arabidopsis thaliana
17 c3h6xA_
100.0
31
PDB header: hydrolaseChain: A: PDB Molecule: dutpase;PDBTitle: crystal structure of dutpase from streptococcus mutans
18 c2okdB_
100.0
29
PDB header: hydrolaseChain: B: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: high resolution crystal structures of vaccinia virus dutpase
19 c3ecyA_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: cg4584-pa, isoform a (bcdna.ld08534);PDBTitle: crystal structural analysis of drosophila melanogaster dutpase
20 c2bazA_
100.0
22
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein bsu20020;PDBTitle: structure of yoss, a putative dutpase from bacillus subtilis
21 d1f7da_
not modelled
100.0
26
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like22 d1duna_
not modelled
100.0
21
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like23 c2d4nA_
not modelled
100.0
35
PDB header: hydrolaseChain: A: PDB Molecule: du;PDBTitle: crystal structure of m-pmv dutpase complexed with dupnpp, substrate2 analogue
24 d2bsya2
not modelled
100.0
20
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like25 c2bt1A_
not modelled
99.9
20
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: epstein barr virus dutpase in complex with a,b-imino dutp
26 c2qxxA_
not modelled
99.9
24
PDB header: hydrolaseChain: A: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: bifunctional dctp deaminase: dutpase from mycobacterium tuberculosis2 in complex with dttp
27 d2bsya1
not modelled
99.9
21
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like28 d1xs1a_
not modelled
99.9
22
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like29 c2qlpC_
not modelled
99.9
26
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: bifunctional dctp deaminase:dutpase from mycobacterium tuberculosis,2 apo form
30 c3km3B_
not modelled
99.9
23
PDB header: hydrolaseChain: B: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: crystal structure of eoxycytidine triphosphate deaminase from2 anaplasma phagocytophilum at 2.1a resolution
31 d1pkha_
not modelled
99.9
25
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like32 c2yzjB_
not modelled
99.9
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: 167aa long hypothetical dutpase;PDBTitle: crystal structure of dctp deaminase from sulfolobus tokodaii
33 c2r9qD_
not modelled
99.8
25
PDB header: hydrolaseChain: D: PDB Molecule: 2'-deoxycytidine 5'-triphosphate deaminase;PDBTitle: crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from2 agrobacterium tumefaciens
34 d1tula_
not modelled
44.2
18
Fold: beta-clipSuperfamily: Tlp20, baculovirus telokin-like proteinFamily: Tlp20, baculovirus telokin-like protein35 d1kwga1
not modelled
17.6
19
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain36 c3kvpB_
not modelled
13.8
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ymzc;PDBTitle: crystal structure of uncharacterized protein ymzc precursor2 from bacillus subtilis, northeast structural genomics3 consortium target sr378a
37 d1ng2a2
not modelled
12.8
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain38 d1v7ra_
not modelled
12.3
38
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)39 c3gzuB_
not modelled
12.2
22
PDB header: virusChain: B: PDB Molecule: inner capsid protein vp2;PDBTitle: vp7 recoated rotavirus dlp
40 c3h0dB_
not modelled
12.1
44
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
41 d1pmia_
not modelled
12.1
14
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Type I phosphomannose isomerase42 c3kz4A_
not modelled
11.5
20
PDB header: virusChain: A: PDB Molecule: inner capsid protein vp2;PDBTitle: crystal structure of the rotavirus double layered particle
43 d1k7ka_
not modelled
10.9
38
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)44 d1dj2a_
not modelled
10.9
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like45 d1dj3a_
not modelled
10.7
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like46 c3tquD_
not modelled
10.7
38
PDB header: hydrolaseChain: D: PDB Molecule: non-canonical purine ntp pyrophosphatase;PDBTitle: structure of a ham1 protein from coxiella burnetii
47 d1qf5a_
not modelled
10.3
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like48 d2cara1
not modelled
9.7
44
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)49 c3ld1A_
not modelled
8.3
27
PDB header: hydrolaseChain: A: PDB Molecule: replicase polyprotein 1a;PDBTitle: crystal structure of ibv nsp2a
50 d1o51a_
not modelled
8.0
67
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: DUF190/COG199351 d1boba_
not modelled
7.5
33
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT52 d3c0na1
not modelled
7.2
18
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Aerolysin/Pertussis toxin (APT) domain53 d1vp2a_
not modelled
6.9
35
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: ITPase (Ham1)54 c1w1fA_
not modelled
6.2
25
PDB header: sh3-domainChain: A: PDB Molecule: tyrosine-protein kinase lyn;PDBTitle: sh3 domain of human lyn tyrosine kinase
55 d1uwfa1
not modelled
6.1
17
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits56 d1x7fa1
not modelled
5.9
31
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Outer surface protein, C-terminal domain57 c3nrfA_
not modelled
5.8
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: apag protein;PDBTitle: crystal structure of an apag protein (pa1934) from pseudomonas2 aeruginosa pao1 at 1.50 a resolution
58 d1e5ba_
not modelled
5.8
6
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family II59 c3d82A_
not modelled
5.6
16
PDB header: metal binding proteinChain: A: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of a cupin-2 domain containing protein (sfri_3543)2 from shewanella frigidimarina ncimb 400 at 2.05 a resolution
60 d1qqfa_
not modelled
5.3
21
Fold: alpha/alpha toroidSuperfamily: Terpenoid cyclases/Protein prenyltransferasesFamily: Complement components61 c2oqbA_
not modelled
5.3
26
PDB header: transferase,gene regulationChain: A: PDB Molecule: histone-arginine methyltransferase carm1;PDBTitle: crystal structure of the n-terminal domain of coactivator-associated2 methyltransferase 1 (carm1)
62 d1cm0a_
not modelled
5.2
18
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT63 c3h1yA_
not modelled
5.2
14
PDB header: isomeraseChain: A: PDB Molecule: mannose-6-phosphate isomerase;PDBTitle: crystal structure of mannose 6-phosphate isomerase from2 salmonella typhimurium bound to substrate (f6p)and metal3 atom (zn)
64 c1xopA_
not modelled
5.1
25
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin;PDBTitle: nmr structure of g1v mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
65 d2ot2a1
not modelled
5.1
29
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like66 c3mwxA_
not modelled
5.1
26
PDB header: isomeraseChain: A: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative galactose mutarotase (bsu18360) from2 bacillus subtilis at 1.45 a resolution