| 1 |
|
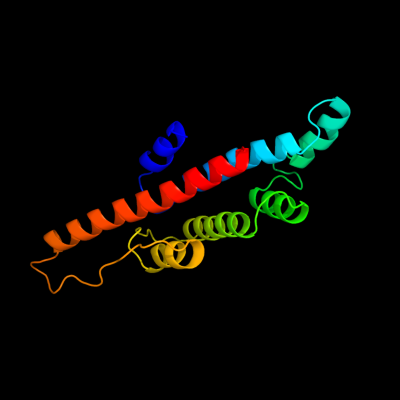
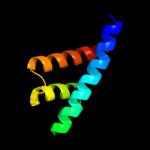
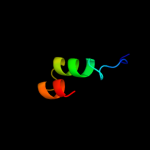
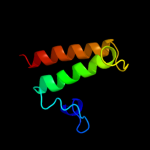
PDB 3brj chain A domain 1
Region: 29 - 189
Aligned: 161
Modelled: 161
Confidence: 100.0%
Identity: 40%
Fold: MtlR-like
Superfamily: MtlR-like
Family: MtlR-like
Phyre2
| 2 |
|
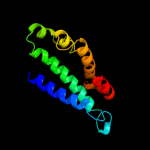
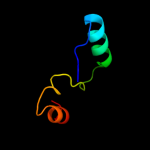
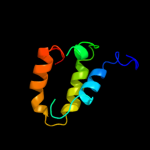
PDB 3c8g chain A domain 1
Region: 29 - 188
Aligned: 153
Modelled: 160
Confidence: 100.0%
Identity: 27%
Fold: MtlR-like
Superfamily: MtlR-like
Family: MtlR-like
Phyre2
| 3 |
|
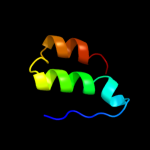
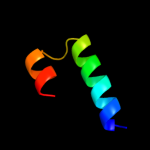
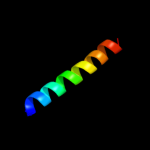
PDB 1wwp chain A
Region: 58 - 122
Aligned: 65
Modelled: 65
Confidence: 59.0%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ttha0636;
PDBTitle: crystal structure of ttk003001694 from thermus thermophilus2 hb8
Phyre2
| 4 |
|
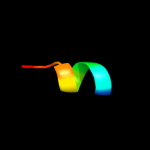
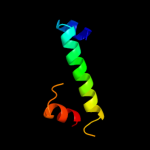
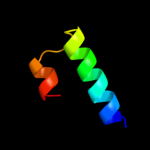
PDB 1wty chain A
Region: 57 - 122
Aligned: 64
Modelled: 66
Confidence: 39.7%
Identity: 13%
Fold: Four-helical up-and-down bundle
Superfamily: Nucleotidyltransferase substrate binding subunit/domain
Family: Family 1 bi-partite nucleotidyltransferase subunit
Phyre2
| 5 |
|
PDB 1ylm chain A
Region: 19 - 140
Aligned: 112
Modelled: 122
Confidence: 21.8%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein bsu32300;
PDBTitle: structure of cytosolic protein of unknown function yute2 from bacillus subtilis
Phyre2
| 6 |
|
PDB 2nn4 chain A domain 1
Region: 93 - 115
Aligned: 23
Modelled: 23
Confidence: 20.1%
Identity: 22%
Fold: YqgQ-like
Superfamily: YqgQ-like
Family: YqgQ-like
Phyre2
| 7 |
|
PDB 2fex chain A domain 1
Region: 78 - 114
Aligned: 36
Modelled: 37
Confidence: 14.9%
Identity: 11%
Fold: Flavodoxin-like
Superfamily: Class I glutamine amidotransferase-like
Family: DJ-1/PfpI
Phyre2
| 8 |
|
PDB 1twf chain C domain 2
Region: 95 - 103
Aligned: 9
Modelled: 9
Confidence: 10.7%
Identity: 33%
Fold: Insert subdomain of RNA polymerase alpha subunit
Superfamily: Insert subdomain of RNA polymerase alpha subunit
Family: Insert subdomain of RNA polymerase alpha subunit
Phyre2
| 9 |
|
PDB 1vkf chain A
Region: 65 - 102
Aligned: 36
Modelled: 38
Confidence: 10.1%
Identity: 11%
Fold: TIM beta/alpha-barrel
Superfamily: GlpP-like
Family: GlpP-like
Phyre2
| 10 |
|
PDB 1gxs chain C
Region: 82 - 110
Aligned: 29
Modelled: 29
Confidence: 10.0%
Identity: 14%
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
Phyre2
| 11 |
|
PDB 1bcr chain A
Region: 84 - 111
Aligned: 28
Modelled: 28
Confidence: 9.7%
Identity: 11%
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
Phyre2
| 12 |
|
PDB 1qki chain A domain 1
Region: 40 - 83
Aligned: 42
Modelled: 44
Confidence: 7.9%
Identity: 17%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 13 |
|
PDB 2gsc chain A domain 1
Region: 84 - 110
Aligned: 27
Modelled: 27
Confidence: 7.3%
Identity: 19%
Fold: Bromodomain-like
Superfamily: IVS-encoded protein-like
Family: IVS-encoded protein-like
Phyre2
| 14 |
|
PDB 1a9x chain A domain 1
Region: 93 - 139
Aligned: 47
Modelled: 47
Confidence: 7.2%
Identity: 23%
Fold: Carbamoyl phosphate synthetase, large subunit connection domain
Superfamily: Carbamoyl phosphate synthetase, large subunit connection domain
Family: Carbamoyl phosphate synthetase, large subunit connection domain
Phyre2
| 15 |
|
PDB 2ztt chain A
Region: 60 - 123
Aligned: 62
Modelled: 64
Confidence: 7.1%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:rna-directed rna polymerase catalytic subunit;
PDBTitle: crystal structure of rna polymerase pb1-pb2 subunits from2 influenza a virus
Phyre2
| 16 |
|
PDB 2ful chain E
Region: 34 - 107
Aligned: 74
Modelled: 74
Confidence: 6.5%
Identity: 19%
PDB header:translation
Chain: E: PDB Molecule:eukaryotic translation initiation factor 5;
PDBTitle: crystal structure of the c-terminal domain of s. cerevisiae eif5
Phyre2
| 17 |
|
PDB 2a45 chain J
Region: 96 - 120
Aligned: 25
Modelled: 25
Confidence: 6.4%
Identity: 16%
PDB header:hydrolase/hydrolase inhibitor
Chain: J: PDB Molecule:fibrinogen alpha chain;
PDBTitle: crystal structure of the complex between thrombin and the central "e"2 region of fibrin
Phyre2
| 18 |
|
PDB 2rld chain A domain 1
Region: 84 - 110
Aligned: 27
Modelled: 27
Confidence: 6.0%
Identity: 22%
Fold: Bromodomain-like
Superfamily: IVS-encoded protein-like
Family: IVS-encoded protein-like
Phyre2
| 19 |
|
PDB 2bcw chain C
Region: 20 - 41
Aligned: 22
Modelled: 22
Confidence: 5.6%
Identity: 9%
PDB header:ribosome
Chain: C: PDB Molecule:elongation factor g;
PDBTitle: coordinates of the n-terminal domain of ribosomal protein2 l11,c-terminal domain of ribosomal protein l7/l12 and a3 portion of the g' domain of elongation factor g, as fitted4 into cryo-em map of an escherichia coli 70s*ef-5 g*gdp*fusidic acid complex
Phyre2
| 20 |
|
PDB 2wpy chain A
Region: 51 - 71
Aligned: 21
Modelled: 21
Confidence: 5.4%
Identity: 19%
PDB header:transcription
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: gcn4 leucine zipper mutant with one vxxnxxx motif2 coordinating chloride
Phyre2
| 21 |
|