| 1 |
|
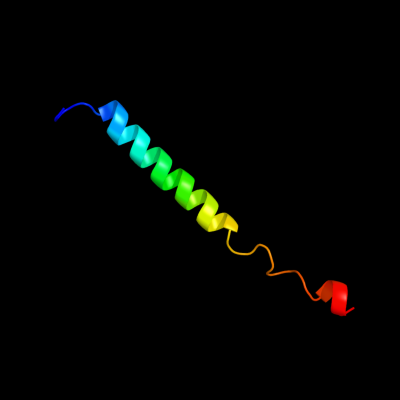
PDB 3caz chain A
Region: 5 - 42
Aligned: 38
Modelled: 38
Confidence: 38.1%
Identity: 32%
PDB header:signaling protein
Chain: A: PDB Molecule:bar protein;
PDBTitle: crystal structure of a bar protein from galdieria sulphuraria
Phyre2
| 2 |
|
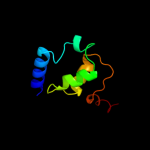
PDB 3sqg chain F
Region: 37 - 50
Aligned: 14
Modelled: 14
Confidence: 28.7%
Identity: 29%
PDB header:transferase
Chain: F: PDB Molecule:methyl-coenzyme m reductase, gamma subunit;
PDBTitle: crystal structure of a methyl-coenzyme m reductase purified from black2 sea mats
Phyre2
| 3 |
|
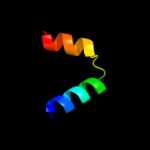
PDB 2yh5 chain A
Region: 37 - 46
Aligned: 10
Modelled: 10
Confidence: 16.9%
Identity: 30%
PDB header:lipid binding protein
Chain: A: PDB Molecule:dapx protein;
PDBTitle: structure of the c-terminal domain of bamc
Phyre2
| 4 |
|
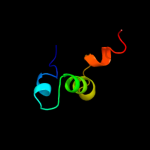
PDB 3gn4 chain A
Region: 72 - 96
Aligned: 25
Modelled: 25
Confidence: 9.8%
Identity: 16%
PDB header:motor protein
Chain: A: PDB Molecule:myosin-vi;
PDBTitle: myosin lever arm
Phyre2
| 5 |
|
PDB 3s6i chain A
Region: 10 - 82
Aligned: 72
Modelled: 73
Confidence: 9.5%
Identity: 14%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:dna-3-methyladenine glycosylase 1;
PDBTitle: schizosaccaromyces pombe 3-methyladenine dna glycosylase (mag1) in2 complex with abasic-dna.
Phyre2
| 6 |
|
PDB 1b0n chain B
Region: 30 - 55
Aligned: 26
Modelled: 26
Confidence: 7.8%
Identity: 19%
PDB header:transcription regulator
Chain: B: PDB Molecule:protein (sini protein);
PDBTitle: sinr protein/sini protein complex
Phyre2
| 7 |
|
PDB 1b0n chain B
Region: 30 - 55
Aligned: 26
Modelled: 26
Confidence: 7.8%
Identity: 19%
Fold: Dimerisation interlock
Superfamily: SinR repressor dimerisation domain-like
Family: SinR repressor dimerisation domain-like
Phyre2
| 8 |
|
PDB 2yg8 chain B
Region: 38 - 80
Aligned: 43
Modelled: 43
Confidence: 7.8%
Identity: 12%
PDB header:hydrolase
Chain: B: PDB Molecule:dna-3-methyladenine glycosidase ii, putative;
PDBTitle: structure of an unusual 3-methyladenine dna glycosylase ii (2 alka) from deinococcus radiodurans
Phyre2