| 1 |
|
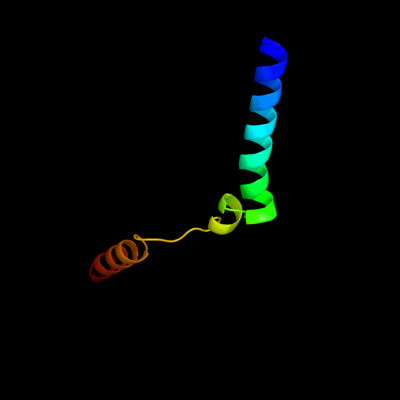
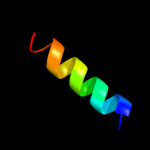
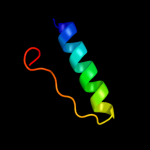
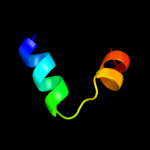
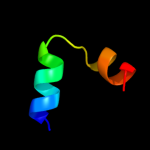
PDB 2oar chain A
Region: 7 - 57
Aligned: 49
Modelled: 51
Confidence: 78.7%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 2 |
|
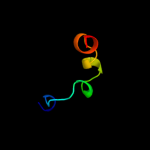
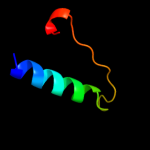
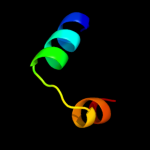
PDB 2rdd chain B
Region: 18 - 58
Aligned: 25
Modelled: 25
Confidence: 54.6%
Identity: 40%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2
| 3 |
|
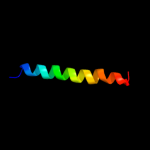
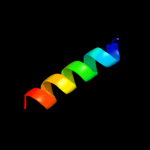
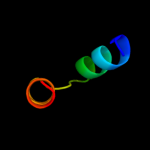
PDB 2ifo chain A
Region: 5 - 35
Aligned: 31
Modelled: 31
Confidence: 39.9%
Identity: 19%
PDB header:virus
Chain: A: PDB Molecule:inovirus;
PDBTitle: model-building studies of inovirus: genetic variations on a2 geometric theme
Phyre2
| 4 |
|
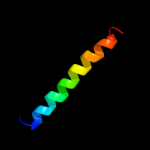
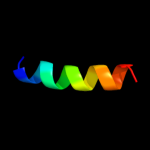
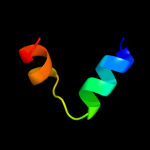
PDB 2oar chain A domain 1
Region: 7 - 49
Aligned: 41
Modelled: 43
Confidence: 35.7%
Identity: 12%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 5 |
|
PDB 3hd7 chain A
Region: 15 - 31
Aligned: 17
Modelled: 17
Confidence: 20.0%
Identity: 35%
PDB header:exocytosis
Chain: A: PDB Molecule:vesicle-associated membrane protein 2;
PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
Phyre2
| 6 |
|
PDB 3lm3 chain A
Region: 70 - 94
Aligned: 25
Modelled: 25
Confidence: 17.5%
Identity: 32%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative glycoside hydrolase/deacetylase2 (bdi_3119) from parabacteroides distasonis at 1.44 a resolution
Phyre2
| 7 |
|
PDB 3fd9 chain C
Region: 55 - 65
Aligned: 11
Modelled: 8
Confidence: 15.2%
Identity: 55%
PDB header:unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the transcriptional anti-activator exsd2 from pseudomonas aeruginosa
Phyre2
| 8 |
|
PDB 2k9y chain B
Region: 7 - 34
Aligned: 28
Modelled: 28
Confidence: 14.1%
Identity: 43%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 9 |
|
PDB 2k9y chain A
Region: 7 - 34
Aligned: 28
Modelled: 28
Confidence: 12.0%
Identity: 43%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 10 |
|
PDB 3l4q chain A
Region: 13 - 44
Aligned: 29
Modelled: 29
Confidence: 11.5%
Identity: 34%
PDB header:viral protein/protein binding
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: structural insights into phosphoinositide 3-kinase2 activation by the influenza a virus ns1 protein
Phyre2
| 11 |
|
PDB 2a93 chain B
Region: 50 - 67
Aligned: 18
Modelled: 18
Confidence: 11.0%
Identity: 50%
PDB header:leucine zippers
Chain: B: PDB Molecule:c-myc-max heterodimeric leucine zipper;
PDBTitle: nmr solution structure of the c-myc-max heterodimeric2 leucine zipper, 40 structures
Phyre2
| 12 |
|
PDB 2gx9 chain A domain 1
Region: 13 - 44
Aligned: 29
Modelled: 32
Confidence: 9.5%
Identity: 34%
Fold: Ns1 effector domain-like
Superfamily: Ns1 effector domain-like
Family: Ns1 effector domain-like
Phyre2
| 13 |
|
PDB 2r6g chain F domain 1
Region: 15 - 34
Aligned: 20
Modelled: 20
Confidence: 8.8%
Identity: 30%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 14 |
|
PDB 2kub chain A
Region: 39 - 55
Aligned: 17
Modelled: 17
Confidence: 6.8%
Identity: 29%
PDB header:structural protein
Chain: A: PDB Molecule:fimbriae-associated protein fap1;
PDBTitle: solution structure of the alpha subdomain of the major non-repeat unit2 of fap1 fimbriae of streptococcus parasanguis
Phyre2
| 15 |
|
PDB 3rgu chain A
Region: 39 - 55
Aligned: 17
Modelled: 17
Confidence: 6.6%
Identity: 29%
PDB header:structural protein
Chain: A: PDB Molecule:fimbriae-associated protein fap1;
PDBTitle: structure of fap-nra at ph 5.0
Phyre2
| 16 |
|
PDB 1f93 chain H
Region: 42 - 63
Aligned: 22
Modelled: 22
Confidence: 6.2%
Identity: 45%
PDB header:transcription
Chain: H: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: crystal structure of a complex between the dimerization2 domain of hnf-1 alpha and the coactivator dcoh
Phyre2
| 17 |
|
PDB 1g39 chain B
Region: 42 - 63
Aligned: 22
Modelled: 22
Confidence: 6.2%
Identity: 45%
PDB header:transcription
Chain: B: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: wild-type hnf-1alpha dimerization domain
Phyre2
| 18 |
|
PDB 1g39 chain D
Region: 42 - 63
Aligned: 22
Modelled: 22
Confidence: 6.2%
Identity: 45%
PDB header:transcription
Chain: D: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: wild-type hnf-1alpha dimerization domain
Phyre2
| 19 |
|
PDB 1f93 chain F
Region: 42 - 63
Aligned: 22
Modelled: 22
Confidence: 6.1%
Identity: 45%
PDB header:transcription
Chain: F: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: crystal structure of a complex between the dimerization2 domain of hnf-1 alpha and the coactivator dcoh
Phyre2
| 20 |
|
PDB 1f93 chain F
Region: 42 - 63
Aligned: 22
Modelled: 22
Confidence: 6.1%
Identity: 45%
Fold: Dimerisation interlock
Superfamily: Dimerization cofactor of HNF-1 alpha
Family: Dimerization cofactor of HNF-1 alpha
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|