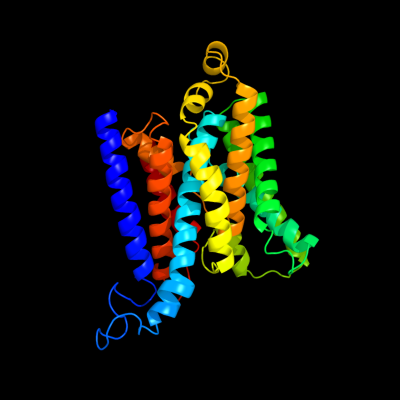
1 d1l7va_
100.0
27
Fold: ABC transporter involved in vitamin B12 uptake, BtuCSuperfamily: ABC transporter involved in vitamin B12 uptake, BtuCFamily: ABC transporter involved in vitamin B12 uptake, BtuC2 c2nq2A_
100.0
27
PDB header: metal transportChain: A: PDB Molecule: hypothetical abc transporter permease proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
3 d1otsa_
51.2
11
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel4 d1kpla_
46.2
14
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel5 c3k3gA_
38.0
11
PDB header: transport proteinChain: A: PDB Molecule: urea transporter;PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
6 c1yceD_
27.6
14
PDB header: membrane proteinChain: D: PDB Molecule: subunit c;PDBTitle: structure of the rotor ring of f-type na+-atpase from ilyobacter2 tartaricus
7 c1q1hA_
22.2
23
PDB header: transcriptionChain: A: PDB Molecule: transcription factor e;PDBTitle: an extended winged helix domain in general transcription2 factor e/iie alpha
8 d1q1ha_
22.2
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcription factor E/IIe-alpha, N-terminal domain9 c2ht2B_
20.9
12
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
10 c2w5jM_
18.6
20
PDB header: hydrolaseChain: M: PDB Molecule: atp synthase c chain, chloroplastic;PDBTitle: structure of the c14-rotor ring of the proton translocating2 chloroplast atp synthase
11 c2xndK_
17.5
21
PDB header: hydrolaseChain: K: PDB Molecule: atp synthase lipid-binding protein, mitochondrial;PDBTitle: crystal structure of bovine f1-c8 sub-complex of atp2 synthase
12 d2p7tc1
16.6
34
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels13 c3lrcC_
15.3
7
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
14 c2b2hA_
15.3
13
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter;PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
15 c2wpdP_
14.5
23
PDB header: hydrolaseChain: P: PDB Molecule: atp synthase subunit 9, mitochondrial;PDBTitle: the mg.adp inhibited state of the yeast f1c10 atp synthase
16 c3qnqD_
14.4
7
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
17 d2r5fa1
13.5
33
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like18 c2kncA_
12.3
32
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
19 c2x2vG_
12.0
18
PDB header: membrane proteinChain: G: PDB Molecule: atp synthase subunit c;PDBTitle: structural basis of a novel proton-coordination type in an2 f1fo-atp synthase rotor ring
20 d1y88a1
11.6
21
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: Hypothetical protein AF1548, C-terminal domain21 c2zt9E_
not modelled
11.5
34
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
22 c2o012_
not modelled
11.2
48
PDB header: photosynthesisChain: 2: PDB Molecule: type ii chlorophyll a/b binding protein fromPDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
23 c3a03A_
not modelled
11.1
11
PDB header: gene regulationChain: A: PDB Molecule: t-cell leukemia homeobox protein 2;PDBTitle: crystal structure of hox11l1 homeodomain
24 c2jd3B_
not modelled
10.8
33
PDB header: dna binding proteinChain: B: PDB Molecule: stbb protein;PDBTitle: parr from plasmid pb171
25 d1du0a_
not modelled
10.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain26 d1x2ma1
not modelled
10.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain27 c3mk7F_
not modelled
10.5
11
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
28 c2kfsA_
not modelled
10.4
28
PDB header: dna-binding proteinChain: A: PDB Molecule: conserved hypothetical regulatory protein;PDBTitle: nmr structure of rv2175c
29 d1ku2a1
not modelled
10.4
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain30 d1rh5b_
not modelled
10.3
33
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit31 c1iijA_
not modelled
10.0
35
PDB header: signaling proteinChain: A: PDB Molecule: erbb-2 receptor protein-tyrosine kinase;PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
32 c2vi6F_
not modelled
9.6
11
PDB header: transcriptionChain: F: PDB Molecule: homeobox protein nanog;PDBTitle: crystal structure of the nanog homeodomain
33 c1l0oC_
not modelled
9.6
28
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
34 d1l0oc_
not modelled
9.6
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain35 c3cmwA_
not modelled
9.5
27
PDB header: recombination/dnaChain: A: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
36 d2p7vb1
not modelled
9.4
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain37 c2da4A_
not modelled
9.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dkfzp686k21156;PDBTitle: solution structure of the homeobox domain of the2 hypothetical protein, dkfzp686k21156
38 c1fftG_
not modelled
9.4
5
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
39 c2xglB_
not modelled
9.3
18
PDB header: antibioticChain: B: PDB Molecule: colicin-m immunity protein;PDBTitle: the x-ray structure of the escherichia coli colicin m immunity2 protein demonstrates the presence of a disulphide bridge, which is3 functionally essential
40 c3nd0A_
not modelled
9.3
10
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
41 c2jwaA_
not modelled
9.3
18
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
42 d1rp3a1
not modelled
9.3
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain43 d1akha_
not modelled
9.2
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain44 d1sana_
not modelled
9.1
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain45 c1yi7C_
not modelled
8.9
31
PDB header: hydrolaseChain: C: PDB Molecule: beta-xylosidase, family 43 glycosyl hydrolase;PDBTitle: beta-d-xylosidase (selenomethionine) xynd from clostridium2 acetobutylicum
46 d1ku3a_
not modelled
8.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain47 c3aqpB_
not modelled
8.8
21
PDB header: membrane proteinChain: B: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
48 c2kz3A_
not modelled
8.8
24
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein rad51l3;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for human rad51d2 from 1 to 83
49 d2o34a1
not modelled
8.7
20
Fold: T-foldSuperfamily: ApbE-likeFamily: DVU1097-like50 c1wu0A_
not modelled
8.5
22
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase c chain;PDBTitle: solution structure of subunit c of f1fo-atp synthase from2 the thermophilic bacillus ps3
51 d1wh5a_
not modelled
8.5
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain52 d1le8a_
not modelled
8.5
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain53 d1ttya_
not modelled
8.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain54 d1p7ia_
not modelled
8.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain55 d2p81a1
not modelled
8.3
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain56 c3t72o_
not modelled
8.3
12
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
57 d2asba2
not modelled
8.2
29
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)58 c3k69A_
not modelled
8.2
19
PDB header: transcriptionChain: A: PDB Molecule: putative transcription regulator;PDBTitle: crystal structure of a putative transcriptional regulator (lp_0360)2 from lactobacillus plantarum at 1.95 a resolution
59 c2da7A_
not modelled
8.2
11
PDB header: dna binding proteinChain: A: PDB Molecule: zinc finger homeobox protein 1b;PDBTitle: solution structure of the homeobox domain of zinc finger2 homeobox protein 1b (smad interacting protein 1)
60 c2xvtC_
not modelled
8.2
42
PDB header: membrane proteinChain: C: PDB Molecule: receptor activity-modifying protein 2;PDBTitle: structure of the extracellular domain of human ramp2
61 c2k1aA_
not modelled
8.2
32
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
62 d1b8ia_
not modelled
8.1
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain63 c2jmlA_
not modelled
8.1
25
PDB header: transcriptionChain: A: PDB Molecule: dna binding domain/transcriptional regulator;PDBTitle: solution structure of the n-terminal domain of cara repressor
64 d1rhzb_
not modelled
8.0
24
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit65 d1mylb_
not modelled
8.0
38
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors66 d1o4xa1
not modelled
7.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain67 d1ku7a_
not modelled
7.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain68 d1rc2a_
not modelled
7.9
26
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like69 c2djnA_
not modelled
7.9
22
PDB header: transcriptionChain: A: PDB Molecule: homeobox protein dlx-5;PDBTitle: the solution structure of the homeobox domain of human2 homeobox protein dlx-5
70 d1bw5a_
not modelled
7.8
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain71 d1f43a_
not modelled
7.8
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain72 c2rn7A_
not modelled
7.7
17
PDB header: unknown functionChain: A: PDB Molecule: is629 orfa;PDBTitle: nmr solution structure of tnpe protein from shigella2 flexneri. northeast structural genomics target sfr125
73 c2l2tA_
not modelled
7.7
24
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
74 c2o8xA_
not modelled
7.7
17
PDB header: transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-c factor;PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
75 c2k40A_
not modelled
7.6
22
PDB header: dna binding proteinChain: A: PDB Molecule: homeobox expressed in es cells 1;PDBTitle: nmr structure of hesx-1 homeodomain double mutant r31l/e42l
76 c3cmuA_
not modelled
7.6
24
PDB header: recombination/dnaChain: A: PDB Molecule: protein reca;PDBTitle: mechanism of homologous recombination from the reca-2 ssdna/dsdna structures
77 d1f6ga_
not modelled
7.6
28
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels78 d1pufa_
not modelled
7.6
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain79 d1s7ea1
not modelled
7.6
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain80 d1b72a_
not modelled
7.6
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain81 c2kv5A_
not modelled
7.6
20
PDB header: toxinChain: A: PDB Molecule: putative uncharacterized protein rnai;PDBTitle: solution structure of the par toxin fst in dpc micelles
82 d2craa1
not modelled
7.5
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain83 d1zq3p1
not modelled
7.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain84 c2x48B_
not modelled
7.5
22
PDB header: viral proteinChain: B: PDB Molecule: cag38821;PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
85 d2hi3a1
not modelled
7.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain86 c3u5eL_
not modelled
7.4
26
PDB header: ribosomeChain: L: PDB Molecule: 60s ribosomal protein l13-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
87 c2wwbB_
not modelled
7.4
28
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sec61 subunit gamma;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
88 c3a01A_
not modelled
7.3
17
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeodomain-containing protein;PDBTitle: crystal structure of aristaless and clawless homeodomains bound to dna
89 d1s9ca1
not modelled
7.3
15
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: MaoC-like90 d1l8qa1
not modelled
7.3
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: Chromosomal replication initiation factor DnaA C-terminal domain IV91 d1js2a_
not modelled
7.3
25
Fold: HIPIP (high potential iron protein)Superfamily: HIPIP (high potential iron protein)Family: HIPIP (high potential iron protein)92 c2yx8A_
not modelled
7.3
25
PDB header: protein transportChain: A: PDB Molecule: receptor activity-modifying protein 1;PDBTitle: crystal structure of the extracellular domain of human ramp1
93 d1ocpa_
not modelled
7.3
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain94 d1myka_
not modelled
7.3
38
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors95 d1smyf2
not modelled
7.2
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain96 c4a18U_
not modelled
7.2
21
PDB header: ribosomeChain: U: PDB Molecule: rpl13;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
97 d1yz8p1
not modelled
7.2
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain98 d2dt5a1
not modelled
7.1
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional repressor Rex, N-terminal domain99 c2dmsA_
not modelled
7.1
22
PDB header: dna binding proteinChain: A: PDB Molecule: homeobox protein otx2;PDBTitle: solution structure of the homeobox domain of homeobox2 protein otx2