| 1 |
|
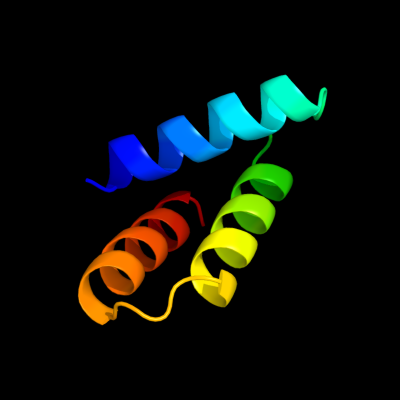
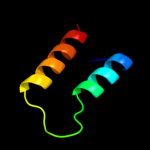
PDB 2oxl chain A
Region: 12 - 64
Aligned: 53
Modelled: 53
Confidence: 72.0%
Identity: 19%
PDB header:gene regulation
Chain: A: PDB Molecule:hypothetical protein ymgb;
PDBTitle: structure and function of the e. coli protein ymgb: a protein critical2 for biofilm formation and acid resistance
Phyre2
| 2 |
|
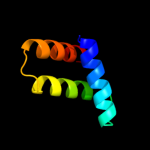
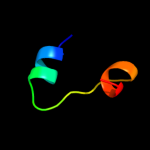
PDB 2bid chain A
Region: 36 - 63
Aligned: 28
Modelled: 28
Confidence: 31.9%
Identity: 25%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 3 |
|
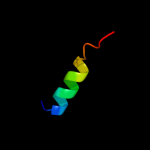
PDB 1ddb chain A
Region: 36 - 63
Aligned: 28
Modelled: 28
Confidence: 30.9%
Identity: 29%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 4 |
|
PDB 1r8j chain A domain 2
Region: 14 - 51
Aligned: 38
Modelled: 38
Confidence: 8.6%
Identity: 13%
Fold: Flavodoxin-like
Superfamily: CheY-like
Family: N-terminal domain of the circadian clock protein KaiA
Phyre2
| 5 |
|
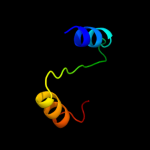
PDB 2uwj chain E
Region: 1 - 26
Aligned: 26
Modelled: 26
Confidence: 8.5%
Identity: 31%
PDB header:chaperone
Chain: E: PDB Molecule:type iii export protein psce;
PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
Phyre2
| 6 |
|
PDB 3bbz chain A
Region: 10 - 60
Aligned: 36
Modelled: 39
Confidence: 7.8%
Identity: 53%
PDB header:viral protein, replication
Chain: A: PDB Molecule:p protein;
PDBTitle: structure of the nucleocapsid-binding domain from the mumps2 virus phosphoprotein
Phyre2
| 7 |
|
PDB 3us8 chain A
Region: 34 - 55
Aligned: 22
Modelled: 21
Confidence: 7.2%
Identity: 50%
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
Phyre2
| 8 |
|
PDB 2a40 chain C
Region: 10 - 28
Aligned: 19
Modelled: 19
Confidence: 6.3%
Identity: 37%
PDB header:structural protein
Chain: C: PDB Molecule:wiskott-aldrich syndrome protein family member 2;
PDBTitle: ternary complex of the wh2 domain of wave with actin-dnase i
Phyre2
| 9 |
|
PDB 2a40 chain F
Region: 10 - 28
Aligned: 19
Modelled: 19
Confidence: 6.3%
Identity: 37%
PDB header:structural protein
Chain: F: PDB Molecule:wiskott-aldrich syndrome protein family member 2;
PDBTitle: ternary complex of the wh2 domain of wave with actin-dnase i
Phyre2