| 1 |
|
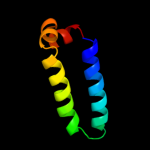
PDB 3p5n chain A
Region: 81 - 112
Aligned: 32
Modelled: 32
Confidence: 69.6%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:riboflavin uptake protein;
PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
Phyre2
| 2 |
|
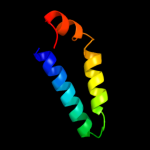
PDB 3qbr chain A
Region: 11 - 66
Aligned: 56
Modelled: 56
Confidence: 39.4%
Identity: 11%
PDB header:apoptosis
Chain: A: PDB Molecule:sjchgc06286 protein;
PDBTitle: bakbh3 in complex with sja
Phyre2
| 3 |
|
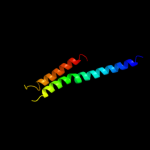
PDB 1f16 chain A
Region: 3 - 66
Aligned: 64
Modelled: 64
Confidence: 36.6%
Identity: 17%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 4 |
|
PDB 1bxl chain A
Region: 11 - 66
Aligned: 56
Modelled: 56
Confidence: 35.4%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 5 |
|
PDB 1ysg chain A domain 1
Region: 11 - 66
Aligned: 56
Modelled: 56
Confidence: 31.1%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 6 |
|
PDB 1o0l chain A
Region: 11 - 66
Aligned: 56
Modelled: 56
Confidence: 29.0%
Identity: 14%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 7 |
|
PDB 2o2f chain A
Region: 11 - 66
Aligned: 56
Modelled: 56
Confidence: 28.3%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:apoptosis regulator bcl-2;
PDBTitle: solution structure of the anti-apoptotic protein bcl-2 in2 complex with an acyl-sulfonamide-based ligand
Phyre2
| 8 |
|
PDB 2pon chain B domain 1
Region: 11 - 69
Aligned: 59
Modelled: 59
Confidence: 26.7%
Identity: 19%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 9 |
|
PDB 3pk1 chain A
Region: 3 - 69
Aligned: 67
Modelled: 67
Confidence: 26.7%
Identity: 12%
PDB header:apoptosis/apoptosis regulator
Chain: A: PDB Molecule:induced myeloid leukemia cell differentiation protein mcl-
PDBTitle: crystal structure of mcl-1 in complex with the baxbh3 domain
Phyre2
| 10 |
|
PDB 1g5m chain A
Region: 11 - 69
Aligned: 59
Modelled: 59
Confidence: 26.3%
Identity: 12%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 11 |
|
PDB 1pq1 chain A
Region: 11 - 69
Aligned: 59
Modelled: 59
Confidence: 25.3%
Identity: 17%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 12 |
|
PDB 2xa0 chain A
Region: 11 - 66
Aligned: 56
Modelled: 56
Confidence: 25.2%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:apoptosis regulator bcl-2;
PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
Phyre2
| 13 |
|
PDB 1zy3 chain A domain 1
Region: 11 - 68
Aligned: 58
Modelled: 58
Confidence: 23.8%
Identity: 16%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 14 |
|
PDB 2jm6 chain B domain 1
Region: 11 - 69
Aligned: 59
Modelled: 59
Confidence: 22.0%
Identity: 15%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 15 |
|
PDB 2yv6 chain A
Region: 11 - 62
Aligned: 52
Modelled: 52
Confidence: 21.8%
Identity: 15%
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2 homologous antagonist/killer;
PDBTitle: crystal structure of human bcl-2 family protein bak
Phyre2
| 16 |
|
PDB 2bbj chain B
Region: 131 - 232
Aligned: 70
Modelled: 71
Confidence: 20.8%
Identity: 23%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2