| 1 |
|
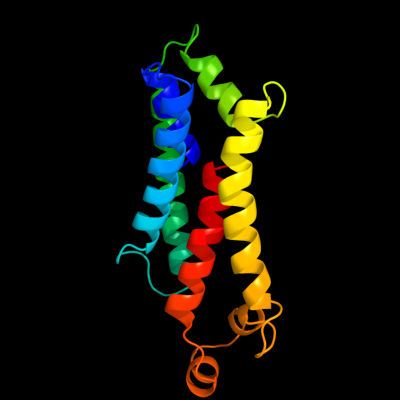
PDB 2nq2 chain A
Region: 149 - 326
Aligned: 148
Modelled: 154
Confidence: 97.7%
Identity: 13%
PDB header:metal transport
Chain: A: PDB Molecule:hypothetical abc transporter permease protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
Phyre2
| 2 |
|
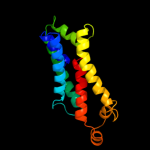
PDB 1l7v chain A
Region: 140 - 326
Aligned: 156
Modelled: 159
Confidence: 96.2%
Identity: 17%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 3 |
|
PDB 1pv7 chain A
Region: 307 - 414
Aligned: 105
Modelled: 108
Confidence: 28.8%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
PDB 2e74 chain G domain 1
Region: 317 - 342
Aligned: 26
Modelled: 26
Confidence: 23.0%
Identity: 27%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 5 |
|
PDB 3pxp chain A
Region: 282 - 314
Aligned: 33
Modelled: 32
Confidence: 22.3%
Identity: 30%
PDB header:transcription regulator
Chain: A: PDB Molecule:helix-turn-helix domain protein;
PDBTitle: crystal structure of a pas and dna binding domain containing protein2 (caur_2278) from chloroflexus aurantiacus j-10-fl at 2.30 a3 resolution
Phyre2
| 6 |
|
PDB 2xut chain C
Region: 313 - 424
Aligned: 112
Modelled: 112
Confidence: 13.9%
Identity: 6%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 7 |
|
PDB 4a18 chain U
Region: 291 - 309
Aligned: 19
Modelled: 19
Confidence: 8.1%
Identity: 16%
PDB header:ribosome
Chain: U: PDB Molecule:rpl13;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
Phyre2
| 8 |
|
PDB 1vf5 chain G
Region: 317 - 336
Aligned: 20
Modelled: 20
Confidence: 7.9%
Identity: 30%
PDB header:photosynthesis
Chain: G: PDB Molecule:protein pet g;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
Phyre2
| 9 |
|
PDB 1vf5 chain G
Region: 317 - 336
Aligned: 20
Modelled: 20
Confidence: 7.9%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 10 |
|
PDB 3u5e chain L
Region: 291 - 309
Aligned: 19
Modelled: 19
Confidence: 7.6%
Identity: 16%
PDB header:ribosome
Chain: L: PDB Molecule:60s ribosomal protein l13-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
Phyre2
| 11 |
|
PDB 1rp1 chain A domain 2
Region: 298 - 327
Aligned: 30
Modelled: 30
Confidence: 7.2%
Identity: 27%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Pancreatic lipase, N-terminal domain
Phyre2
| 12 |
|
PDB 1y9q chain A
Region: 284 - 314
Aligned: 31
Modelled: 26
Confidence: 6.9%
Identity: 13%
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, hth_3 family;
PDBTitle: crystal structure of hth_3 family transcriptional regulator2 from vibrio cholerae
Phyre2
| 13 |
|
PDB 1y6u chain A
Region: 298 - 315
Aligned: 18
Modelled: 18
Confidence: 5.7%
Identity: 17%
PDB header:dna binding protein
Chain: A: PDB Molecule:excisionase from transposon tn916;
PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
Phyre2
| 14 |
|
PDB 1hpl chain A domain 2
Region: 298 - 327
Aligned: 30
Modelled: 30
Confidence: 5.5%
Identity: 23%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Pancreatic lipase, N-terminal domain
Phyre2
| 15 |
|
PDB 1eth chain A domain 2
Region: 298 - 327
Aligned: 30
Modelled: 30
Confidence: 5.2%
Identity: 33%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Pancreatic lipase, N-terminal domain
Phyre2