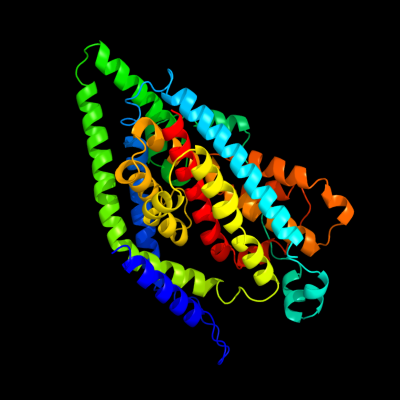
| 1 | d2nwwa1
|
|
 |
100.0 |
30 |
Fold:Proton glutamate symport protein
Superfamily:Proton glutamate symport protein
Family:Proton glutamate symport protein |
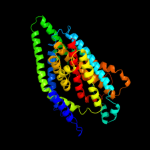
| 2 | c3qnqD_
|
|
 |
54.9 |
30 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
| 3 | d1a77a1
|
|
 |
18.8 |
11 |
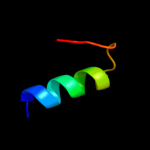
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 4 | c2yvxD_
|
|
 |
16.1 |
20 |
PDB header:transport protein
Chain: D: PDB Molecule:mg2+ transporter mgte;
PDBTitle: crystal structure of magnesium transporter mgte
|
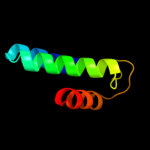
| 5 | c1x0lB_
|
|
 |
14.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
| 6 | d1o26a_
|
|
 |
13.4 |
19 |
Fold:Thymidylate synthase-complementing protein Thy1
Superfamily:Thymidylate synthase-complementing protein Thy1
Family:Thymidylate synthase-complementing protein Thy1 |
| 7 | c3ah5E_
|
|
 |
13.0 |
9 |
PDB header:transferase
Chain: E: PDB Molecule:thymidylate synthase thyx;
PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
|
| 8 | c3blxM_
|
|
 |
8.7 |
17 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 9 | d1ul1x1
|
|
 |
8.2 |
14 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 10 | d1rxwa1
|
|
 |
7.9 |
22 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 11 | c2cfaB_
|
|
 |
7.2 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:thymidylate synthase;
PDBTitle: structure of viral flavin-dependant thymidylate synthase2 thyx
|
| 12 | d1mc8a1
|
|
 |
7.1 |
18 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 13 | d2pxrc1
|
|
 |
7.0 |
16 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 14 | d1m9dc_
|
|
 |
6.9 |
16 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 15 | d1m9fd_
|
|
 |
6.5 |
16 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 16 | c1vf5E_
|
|
 |
6.4 |
22 |
PDB header:photosynthesis
Chain: E: PDB Molecule:protein pet l;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
|
| 17 | c2e75E_
|
|
 |
6.4 |
22 |
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
|
| 18 | c1vf5R_
|
|
 |
6.4 |
22 |
PDB header:photosynthesis
Chain: R: PDB Molecule:protein pet l;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
|
| 19 | c2e76E_
|
|
 |
6.4 |
22 |
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
| 20 | c2e74E_
|
|
 |
6.4 |
22 |
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
|
| 21 | d2e74e1 |
|
not modelled |
6.4 |
22 |
Fold:Single transmembrane helix
Superfamily:PetL subunit of the cytochrome b6f complex
Family:PetL subunit of the cytochrome b6f complex |
| 22 | c2wlvA_ |
|
not modelled |
6.3 |
16 |
PDB header:virus protein
Chain: A: PDB Molecule:gag polyprotein;
PDBTitle: structure of the n-terminal capsid domain of hiv-2
|
| 23 | c2jwaA_ |
|
not modelled |
6.2 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
|
| 24 | d1b43a1 |
|
not modelled |
6.1 |
18 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 25 | d1hqsa_ |
|
not modelled |
6.1 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 26 | d1xo1a1 |
|
not modelled |
6.1 |
18 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 27 | d1ku2a1 |
|
not modelled |
5.9 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
| 28 | d1uxca_ |
|
not modelled |
5.8 |
24 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:GalR/LacI-like bacterial regulator |