| 1 |
|
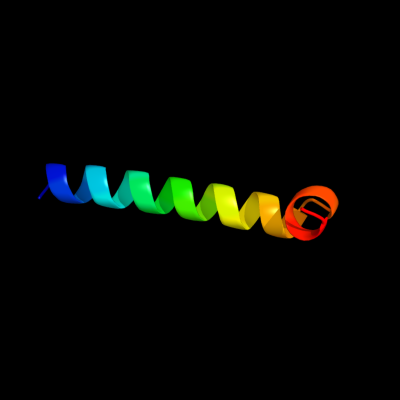
PDB 1nkz chain A
Region: 477 - 507
Aligned: 31
Modelled: 31
Confidence: 37.5%
Identity: 23%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 2 |
|
PDB 1ijd chain A
Region: 477 - 507
Aligned: 31
Modelled: 31
Confidence: 37.3%
Identity: 23%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 3 |
|
PDB 1j4n chain A
Region: 190 - 278
Aligned: 83
Modelled: 89
Confidence: 31.0%
Identity: 17%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 4 |
|
PDB 1ppj chain C domain 1
Region: 117 - 246
Aligned: 91
Modelled: 91
Confidence: 11.1%
Identity: 20%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 5 |
|
PDB 3cx5 chain C domain 1
Region: 117 - 246
Aligned: 91
Modelled: 91
Confidence: 9.8%
Identity: 20%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 6 |
|
PDB 1f6g chain A
Region: 429 - 506
Aligned: 54
Modelled: 54
Confidence: 8.9%
Identity: 17%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 7 |
|
PDB 2knc chain A
Region: 246 - 278
Aligned: 33
Modelled: 33
Confidence: 7.2%
Identity: 18%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 8 |
|
PDB 3d9s chain B
Region: 190 - 285
Aligned: 90
Modelled: 96
Confidence: 6.3%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin-5;
PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
Phyre2
| 9 |
|
PDB 1bcc chain C domain 2
Region: 117 - 246
Aligned: 91
Modelled: 91
Confidence: 6.1%
Identity: 21%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 10 |
|
PDB 2dxb chain R
Region: 11 - 22
Aligned: 12
Modelled: 12
Confidence: 5.9%
Identity: 67%
PDB header:hydrolase
Chain: R: PDB Molecule:thiocyanate hydrolase subunit gamma;
PDBTitle: recombinant thiocyanate hydrolase comprising partially-modified cobalt2 centers
Phyre2
| 11 |
|
PDB 3mk7 chain F
Region: 239 - 311
Aligned: 73
Modelled: 73
Confidence: 5.6%
Identity: 8%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 12 |
|
PDB 1rc2 chain A
Region: 191 - 269
Aligned: 79
Modelled: 79
Confidence: 5.4%
Identity: 10%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 13 |
|
PDB 2oar chain A domain 1
Region: 200 - 282
Aligned: 83
Modelled: 83
Confidence: 5.4%
Identity: 18%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 14 |
|
PDB 3aag chain A
Region: 461 - 474
Aligned: 14
Modelled: 14
Confidence: 5.4%
Identity: 7%
PDB header:transferase
Chain: A: PDB Molecule:general glycosylation pathway protein;
PDBTitle: crystal structure of c. jejuni pglb c-terminal domain
Phyre2