| 1 |
|
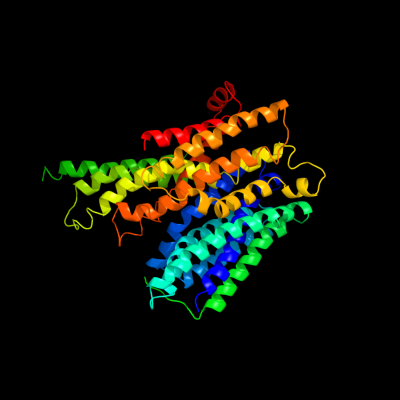
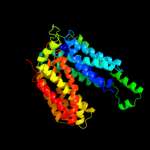
PDB 2xut chain C
Region: 22 - 476
Aligned: 448
Modelled: 455
Confidence: 100.0%
Identity: 21%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 2 |
|
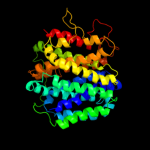
PDB 1pw4 chain A
Region: 1 - 492
Aligned: 423
Modelled: 426
Confidence: 100.0%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
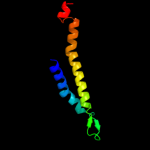
PDB 1pv7 chain A
Region: 21 - 494
Aligned: 405
Modelled: 416
Confidence: 100.0%
Identity: 12%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
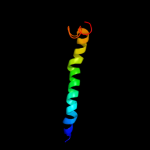
PDB 3o7p chain A
Region: 24 - 482
Aligned: 406
Modelled: 411
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2gfp chain A
Region: 23 - 473
Aligned: 369
Modelled: 396
Confidence: 100.0%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 461 - 496
Aligned: 36
Modelled: 36
Confidence: 70.4%
Identity: 17%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 2oar chain A domain 1
Region: 418 - 499
Aligned: 82
Modelled: 82
Confidence: 51.4%
Identity: 7%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 8 |
|
PDB 2jp3 chain A
Region: 452 - 498
Aligned: 46
Modelled: 47
Confidence: 16.8%
Identity: 13%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 9 |
|
PDB 2oar chain A
Region: 418 - 499
Aligned: 82
Modelled: 82
Confidence: 16.2%
Identity: 7%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 462 - 496
Aligned: 35
Modelled: 35
Confidence: 12.6%
Identity: 17%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 11 |
|
PDB 3hzq chain A
Region: 418 - 489
Aligned: 72
Modelled: 72
Confidence: 11.4%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
Phyre2
| 12 |
|
PDB 2jln chain A
Region: 460 - 498
Aligned: 39
Modelled: 39
Confidence: 10.1%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 13 |
|
PDB 2b2h chain A
Region: 423 - 497
Aligned: 75
Modelled: 75
Confidence: 8.8%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 14 |
|
PDB 3pro chain C domain 1
Region: 42 - 86
Aligned: 45
Modelled: 45
Confidence: 7.0%
Identity: 13%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2
| 15 |
|
PDB 1jo5 chain A
Region: 455 - 482
Aligned: 28
Modelled: 28
Confidence: 6.5%
Identity: 14%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2