| 1 |
|
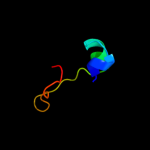
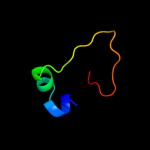
PDB 1jal chain A domain 2
Region: 40 - 48
Aligned: 9
Modelled: 9
Confidence: 39.1%
Identity: 67%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: TGS-like
Family: G domain-linked domain
Phyre2
| 2 |
|
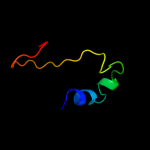
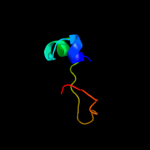
PDB 1ni3 chain A domain 2
Region: 16 - 48
Aligned: 33
Modelled: 33
Confidence: 38.6%
Identity: 18%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: TGS-like
Family: G domain-linked domain
Phyre2
| 3 |
|
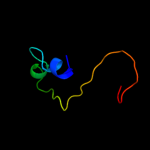
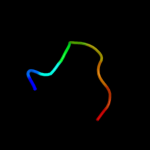
PDB 2hj1 chain A
Region: 5 - 46
Aligned: 40
Modelled: 42
Confidence: 36.2%
Identity: 33%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a 3d domain-swapped dimer of protein hi0395 from2 haemophilus influenzae
Phyre2
| 4 |
|
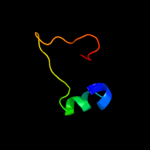
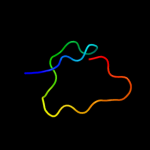
PDB 2hj1 chain A domain 1
Region: 5 - 46
Aligned: 40
Modelled: 42
Confidence: 36.2%
Identity: 33%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: MoaD/ThiS
Family: HI0395-like
Phyre2
| 5 |
|
PDB 1ni3 chain A
Region: 16 - 48
Aligned: 33
Modelled: 33
Confidence: 25.6%
Identity: 18%
PDB header:hydrolase
Chain: A: PDB Molecule:ychf gtp-binding protein;
PDBTitle: structure of the schizosaccharomyces pombe ychf gtpase
Phyre2
| 6 |
|
PDB 2ohf chain A
Region: 16 - 48
Aligned: 33
Modelled: 33
Confidence: 25.3%
Identity: 24%
PDB header:hydrolase
Chain: A: PDB Molecule:gtp-binding protein 9;
PDBTitle: crystal structure of human ola1 in complex with amppcp
Phyre2
| 7 |
|
PDB 1jal chain A
Region: 16 - 48
Aligned: 33
Modelled: 33
Confidence: 21.4%
Identity: 33%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ychf protein;
PDBTitle: ychf protein (hi0393)
Phyre2
| 8 |
|
PDB 2dwq chain B
Region: 40 - 48
Aligned: 9
Modelled: 9
Confidence: 18.7%
Identity: 56%
PDB header:hydrolase
Chain: B: PDB Molecule:gtp-binding protein;
PDBTitle: thermus thermophilus ychf gtp-binding protein
Phyre2
| 9 |
|
PDB 2i9c chain A domain 1
Region: 93 - 106
Aligned: 14
Modelled: 13
Confidence: 11.9%
Identity: 29%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: RPA1889-like
Phyre2
| 10 |
|
PDB 1hrt chain I
Region: 54 - 83
Aligned: 30
Modelled: 30
Confidence: 9.3%
Identity: 33%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Leech antihemostatic proteins
Family: Hirudin-like
Phyre2
| 11 |
|
PDB 1wxq chain A
Region: 25 - 48
Aligned: 24
Modelled: 24
Confidence: 9.1%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:gtp-binding protein;
PDBTitle: crystal structure of gtp binding protein from pyrococcus horikoshii2 ot3
Phyre2
| 12 |
|
PDB 3b49 chain A
Region: 62 - 115
Aligned: 54
Modelled: 54
Confidence: 8.0%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2189 protein;
PDBTitle: crystal structure of an uncharacterized conserved protein from2 listeria innocua
Phyre2
| 13 |
|
PDB 3cs5 chain B
Region: 33 - 47
Aligned: 15
Modelled: 15
Confidence: 8.0%
Identity: 60%
PDB header:photosynthesis
Chain: B: PDB Molecule:phycobilisome degradation protein nbla;
PDBTitle: nbla protein from synechococcus elongatus pcc 7942
Phyre2
| 14 |
|
PDB 3rh3 chain A
Region: 4 - 23
Aligned: 20
Modelled: 20
Confidence: 7.3%
Identity: 35%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized duf3829-like protein;
PDBTitle: crystal structure of an uncharacterized duf3829-like protein (bt_1908)2 from bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution
Phyre2
| 15 |
|
PDB 1nh2 chain B
Region: 7 - 26
Aligned: 18
Modelled: 20
Confidence: 7.1%
Identity: 39%
Fold: Transcription factor IIA (TFIIA), alpha-helical domain
Superfamily: Transcription factor IIA (TFIIA), alpha-helical domain
Family: Transcription factor IIA (TFIIA), alpha-helical domain
Phyre2
| 16 |
|
PDB 3d6n chain B
Region: 63 - 96
Aligned: 30
Modelled: 34
Confidence: 6.6%
Identity: 17%
PDB header:hydrolase/transferase
Chain: B: PDB Molecule:aspartate carbamoyltransferase;
PDBTitle: crystal structure of aquifex dihydroorotase activated by aspartate2 transcarbamoylase
Phyre2
| 17 |
|
PDB 2kd3 chain A
Region: 60 - 66
Aligned: 7
Modelled: 7
Confidence: 6.6%
Identity: 43%
PDB header:protein binding
Chain: A: PDB Molecule:sclerostin;
PDBTitle: nmr structure of the wnt modulator protein sclerostin
Phyre2
| 18 |
|
PDB 2iay chain A domain 1
Region: 101 - 118
Aligned: 18
Modelled: 18
Confidence: 6.6%
Identity: 39%
Fold: Lp2179-like
Superfamily: Lp2179-like
Family: Lp2179-like
Phyre2
| 19 |
|
PDB 2qqr chain A domain 2
Region: 39 - 53
Aligned: 15
Modelled: 15
Confidence: 6.2%
Identity: 40%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: Tudor domain
Phyre2
| 20 |
|
PDB 2qqs chain A domain 2
Region: 39 - 53
Aligned: 15
Modelled: 15
Confidence: 6.1%
Identity: 47%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: Tudor domain
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|