| 1 |
|
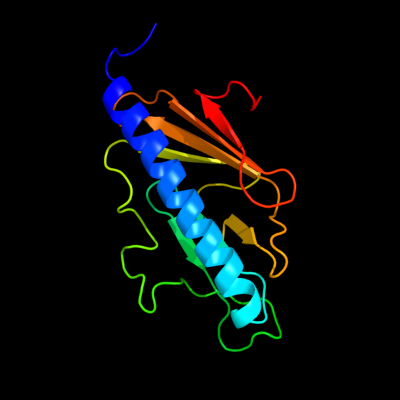
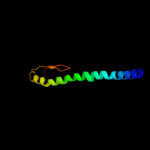
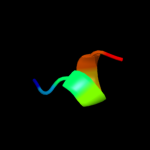
PDB 2knq chain A
Region: 28 - 169
Aligned: 142
Modelled: 142
Confidence: 99.9%
Identity: 98%
PDB header:protein transport
Chain: A: PDB Molecule:general secretion pathway protein h;
PDBTitle: solution structure of e.coli gsph
Phyre2
| 2 |
|
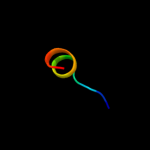
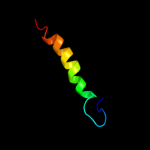
PDB 2qv8 chain B
Region: 39 - 160
Aligned: 116
Modelled: 122
Confidence: 99.5%
Identity: 22%
PDB header:transport protein
Chain: B: PDB Molecule:general secretion pathway protein h;
PDBTitle: structure of the minor pseudopilin epsh from the type 2 secretion2 system of vibrio cholerae
Phyre2
| 3 |
|
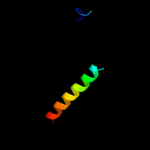
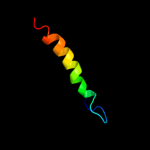
PDB 1oqw chain A
Region: 7 - 156
Aligned: 132
Modelled: 134
Confidence: 99.4%
Identity: 17%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 4 |
|
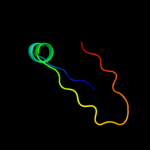
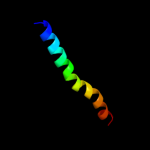
PDB 3sok chain B
Region: 7 - 86
Aligned: 79
Modelled: 80
Confidence: 99.4%
Identity: 20%
PDB header:cell adhesion
Chain: B: PDB Molecule:fimbrial protein;
PDBTitle: dichelobacter nodosus pilin fima
Phyre2
| 5 |
|
PDB 2pil chain A
Region: 8 - 65
Aligned: 56
Modelled: 58
Confidence: 99.2%
Identity: 27%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 6 |
|
PDB 4a18 chain U
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 34.7%
Identity: 55%
PDB header:ribosome
Chain: U: PDB Molecule:rpl13;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
Phyre2
| 7 |
|
PDB 3u5e chain L
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 33.1%
Identity: 55%
PDB header:ribosome
Chain: L: PDB Molecule:60s ribosomal protein l13-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
Phyre2
| 8 |
|
PDB 3dtu chain B domain 2
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 15.9%
Identity: 21%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 9 |
|
PDB 2ii8 chain F
Region: 123 - 152
Aligned: 30
Modelled: 30
Confidence: 14.5%
Identity: 23%
PDB header:signaling protein
Chain: F: PDB Molecule:anabaena sensory rhodopsin transducer protein;
PDBTitle: anabaena sensory rhodopsin transducer
Phyre2
| 10 |
|
PDB 1v54 chain B domain 2
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 14.4%
Identity: 10%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 11 |
|
PDB 3ehb chain B domain 2
Region: 8 - 30
Aligned: 23
Modelled: 23
Confidence: 12.6%
Identity: 26%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 2wsf chain G
Region: 5 - 13
Aligned: 9
Modelled: 9
Confidence: 11.5%
Identity: 22%
PDB header:photosynthesis
Chain: G: PDB Molecule:photosystem i reaction center subunit v,
PDBTitle: improved model of plant photosystem i
Phyre2
| 13 |
|
PDB 2kxe chain A
Region: 7 - 13
Aligned: 7
Modelled: 7
Confidence: 9.7%
Identity: 43%
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase ii small subunit;
PDBTitle: n-terminal domain of the dp1 subunit of an archaeal d-family dna2 polymerase
Phyre2
| 14 |
|
PDB 1jb0 chain K
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 9.6%
Identity: 17%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem 1 reaction centre subunit x;
PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
Phyre2
| 15 |
|
PDB 1jb0 chain K
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 9.6%
Identity: 17%
Fold: Photosystem I reaction center subunit X, PsaK
Superfamily: Photosystem I reaction center subunit X, PsaK
Family: Photosystem I reaction center subunit X, PsaK
Phyre2
| 16 |
|
PDB 1afo chain B
Region: 11 - 25
Aligned: 15
Modelled: 15
Confidence: 9.4%
Identity: 27%
PDB header:integral membrane protein
Chain: B: PDB Molecule:glycophorin a;
PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
Phyre2
| 17 |
|
PDB 1qle chain B
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 8.6%
Identity: 21%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
Phyre2
| 18 |
|
PDB 1ar1 chain B
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 8.6%
Identity: 21%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2
| 19 |
|
PDB 2kb1 chain A
Region: 7 - 38
Aligned: 32
Modelled: 32
Confidence: 8.2%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2
| 20 |
|
PDB 3lw5 chain K
Region: 5 - 32
Aligned: 28
Modelled: 28
Confidence: 7.4%
Identity: 21%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit x psak;
PDBTitle: improved model of plant photosystem i
Phyre2
| 21 |
|