| 1 |
|
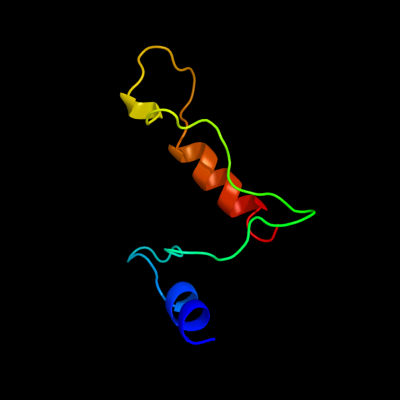
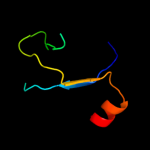
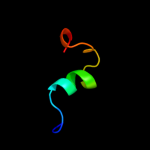
PDB 2v4j chain E
Region: 57 - 118
Aligned: 61
Modelled: 62
Confidence: 36.6%
Identity: 18%
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
Phyre2
| 2 |
|
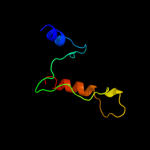
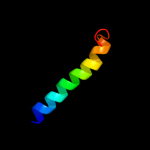
PDB 3c07 chain A domain 2
Region: 37 - 65
Aligned: 29
Modelled: 27
Confidence: 31.2%
Identity: 17%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 3 |
|
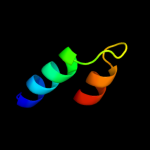
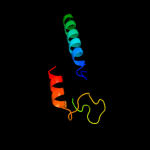
PDB 2jso chain A
Region: 145 - 188
Aligned: 44
Modelled: 44
Confidence: 18.8%
Identity: 23%
PDB header:signaling protein
Chain: A: PDB Molecule:polymyxin resistance protein pmrd;
PDBTitle: antimicrobial resistance protein
Phyre2
| 4 |
|
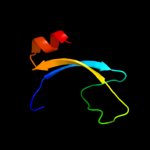
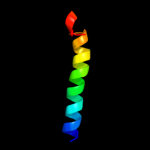
PDB 2rqx chain A
Region: 145 - 188
Aligned: 44
Modelled: 44
Confidence: 13.1%
Identity: 23%
PDB header:signaling protein
Chain: A: PDB Molecule:polymyxin b resistance protein;
PDBTitle: solution nmr structure of pmrd from klebsiella pneumoniae
Phyre2
| 5 |
|
PDB 1oed chain A
Region: 29 - 58
Aligned: 30
Modelled: 30
Confidence: 12.2%
Identity: 17%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 6 |
|
PDB 1qjw chain A
Region: 49 - 113
Aligned: 56
Modelled: 56
Confidence: 9.7%
Identity: 21%
Fold: 7-stranded beta/alpha barrel
Superfamily: Glycosyl hydrolases family 6, cellulases
Family: Glycosyl hydrolases family 6, cellulases
Phyre2
| 7 |
|
PDB 1oed chain E
Region: 29 - 59
Aligned: 31
Modelled: 31
Confidence: 8.5%
Identity: 13%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 8 |
|
PDB 3kdp chain G
Region: 127 - 138
Aligned: 12
Modelled: 12
Confidence: 7.2%
Identity: 42%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 9 |
|
PDB 3kdp chain H
Region: 127 - 138
Aligned: 12
Modelled: 12
Confidence: 7.2%
Identity: 42%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 10 |
|
PDB 2bid chain A
Region: 57 - 101
Aligned: 44
Modelled: 45
Confidence: 7.0%
Identity: 11%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 11 |
|
PDB 1doa chain B
Region: 142 - 169
Aligned: 22
Modelled: 28
Confidence: 6.6%
Identity: 32%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: RhoGDI-like
Phyre2
| 12 |
|
PDB 2k58 chain B
Region: 34 - 50
Aligned: 17
Modelled: 17
Confidence: 6.6%
Identity: 24%
PDB header:transport protein
Chain: B: PDB Molecule:neuronal acetylcholine receptor subunit beta-2;
PDBTitle: nmr structures of the first transmembrane domain of the2 neuronal acetylcholine receptor beta 2 subunit
Phyre2
| 13 |
|
PDB 3hei chain I
Region: 74 - 88
Aligned: 15
Modelled: 15
Confidence: 6.5%
Identity: 40%
PDB header:transferase/signaling protein
Chain: I: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: ligand recognition by a-class eph receptors: crystal structures of the2 epha2 ligand-binding domain and the epha2/ephrin-a1 complex
Phyre2
| 14 |
|
PDB 1oey chain J
Region: 57 - 78
Aligned: 20
Modelled: 19
Confidence: 5.8%
Identity: 20%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 15 |
|
PDB 2gp4 chain A domain 2
Region: 51 - 60
Aligned: 10
Modelled: 10
Confidence: 5.7%
Identity: 40%
Fold: IlvD/EDD N-terminal domain-like
Superfamily: IlvD/EDD N-terminal domain-like
Family: lvD/EDD N-terminal domain-like
Phyre2
| 16 |
|
PDB 2bod chain X domain 1
Region: 49 - 113
Aligned: 46
Modelled: 46
Confidence: 5.3%
Identity: 35%
Fold: 7-stranded beta/alpha barrel
Superfamily: Glycosyl hydrolases family 6, cellulases
Family: Glycosyl hydrolases family 6, cellulases
Phyre2
| 17 |
|
PDB 1dys chain A
Region: 49 - 116
Aligned: 63
Modelled: 68
Confidence: 5.3%
Identity: 24%
Fold: 7-stranded beta/alpha barrel
Superfamily: Glycosyl hydrolases family 6, cellulases
Family: Glycosyl hydrolases family 6, cellulases
Phyre2
| 18 |
|
PDB 2ytu chain A
Region: 21 - 30
Aligned: 10
Modelled: 10
Confidence: 5.2%
Identity: 40%
PDB header:signaling protein
Chain: A: PDB Molecule:friend leukemia integration 1 transcription
PDBTitle: solution structure of the sam_pnt-domain of the human2 friend leukemiaintegration 1 transcription factor
Phyre2
| 19 |
|
PDB 3bz6 chain A domain 2
Region: 36 - 58
Aligned: 22
Modelled: 23
Confidence: 5.2%
Identity: 27%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: PSPTO2686-like
Phyre2
| 20 |
|
PDB 2kw3 chain C
Region: 137 - 144
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 50%
PDB header:dna binding protein
Chain: C: PDB Molecule:regulatory factor x-associated protein;
PDBTitle: heterotrimeric interaction between rfx5 and rfxap
Phyre2
| 21 |
|