| 1 |
|
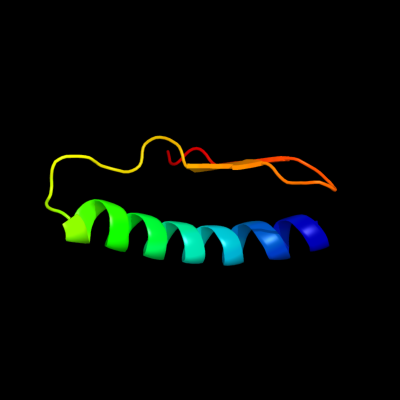
PDB 1y4o chain A domain 1
Region: 18 - 65
Aligned: 48
Modelled: 48
Confidence: 27.0%
Identity: 17%
Fold: Profilin-like
Superfamily: Roadblock/LC7 domain
Family: Roadblock/LC7 domain
Phyre2
| 2 |
|
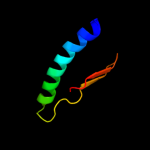
PDB 2hz5 chain A domain 1
Region: 18 - 62
Aligned: 45
Modelled: 45
Confidence: 25.7%
Identity: 18%
Fold: Profilin-like
Superfamily: Roadblock/LC7 domain
Family: Roadblock/LC7 domain
Phyre2
| 3 |
|
PDB 1prt chain B domain 1
Region: 44 - 60
Aligned: 17
Modelled: 17
Confidence: 11.8%
Identity: 41%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Bacterial AB5 toxins, B-subunits
Phyre2
| 4 |
|
PDB 1prt chain C domain 1
Region: 44 - 60
Aligned: 17
Modelled: 17
Confidence: 9.2%
Identity: 41%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Bacterial AB5 toxins, B-subunits
Phyre2
| 5 |
|
PDB 1bcp chain H
Region: 44 - 60
Aligned: 17
Modelled: 17
Confidence: 7.1%
Identity: 41%
PDB header:toxin
Chain: H: PDB Molecule:pertussis toxin;
PDBTitle: binary complex of pertussis toxin and atp
Phyre2
| 6 |
|
PDB 1u7m chain B
Region: 16 - 24
Aligned: 9
Modelled: 9
Confidence: 7.1%
Identity: 44%
PDB header:de novo protein
Chain: B: PDB Molecule:four-helix bundle model;
PDBTitle: solution structure of a diiron protein model: due ferri(ii)2 turn mutant
Phyre2
| 7 |
|
PDB 2jug chain B
Region: 5 - 31
Aligned: 27
Modelled: 27
Confidence: 6.9%
Identity: 11%
PDB header:biosynthetic protein
Chain: B: PDB Molecule:tubc protein;
PDBTitle: multienzyme docking in hybrid megasynthetases
Phyre2
| 8 |
|
PDB 2rpi chain A
Region: 35 - 55
Aligned: 21
Modelled: 21
Confidence: 6.6%
Identity: 29%
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease h;
PDBTitle: the nmr structure of the submillisecond folding2 intermediate of the thermus thermophilus ribonuclease h
Phyre2
| 9 |
|
PDB 1zll chain E
Region: 6 - 38
Aligned: 33
Modelled: 33
Confidence: 5.9%
Identity: 24%
PDB header:membrane protein/signaling protein
Chain: E: PDB Molecule:cardiac phospholamban;
PDBTitle: nmr structure of unphosphorylated human phospholamban2 pentamer
Phyre2
| 10 |
|
PDB 1ehs chain A
Region: 45 - 50
Aligned: 6
Modelled: 6
Confidence: 5.4%
Identity: 83%
Fold: Toxic hairpin
Superfamily: Heat-stable enterotoxin B
Family: Heat-stable enterotoxin B
Phyre2