| 1 |
|
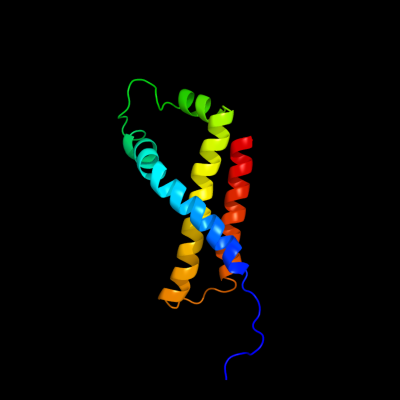
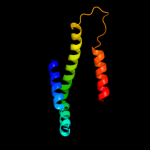
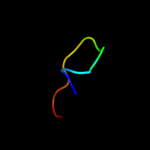
PDB 1kf6 chain D
Region: 1 - 119
Aligned: 119
Modelled: 119
Confidence: 100.0%
Identity: 100%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 2 |
|
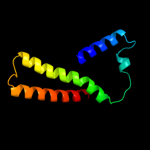
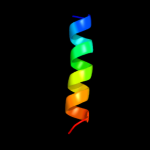
PDB 1nek chain D
Region: 28 - 119
Aligned: 91
Modelled: 92
Confidence: 96.1%
Identity: 19%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 3 |
|
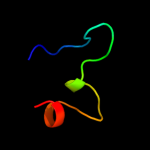
PDB 1nek chain C
Region: 1 - 116
Aligned: 114
Modelled: 116
Confidence: 96.1%
Identity: 15%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 4 |
|
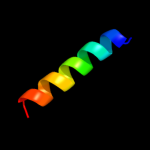
PDB 1kf6 chain C
Region: 28 - 117
Aligned: 90
Modelled: 90
Confidence: 74.3%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 5 |
|
PDB 1yq3 chain C
Region: 32 - 119
Aligned: 88
Modelled: 88
Confidence: 26.7%
Identity: 10%
PDB header:oxidoreductase
Chain: C: PDB Molecule:succinate dehydrogenase cytochrome b, large subunit;
PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
Phyre2
| 6 |
|
PDB 1v54 chain M
Region: 19 - 54
Aligned: 32
Modelled: 36
Confidence: 23.7%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Family: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Phyre2
| 7 |
|
PDB 3kdp chain H
Region: 94 - 113
Aligned: 20
Modelled: 20
Confidence: 18.5%
Identity: 35%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 8 |
|
PDB 3kdp chain G
Region: 94 - 113
Aligned: 20
Modelled: 20
Confidence: 18.5%
Identity: 35%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 9 |
|
PDB 2y69 chain Z
Region: 19 - 54
Aligned: 32
Modelled: 36
Confidence: 16.6%
Identity: 16%
PDB header:electron transport
Chain: Z: PDB Molecule:cytochrome c oxidase polypeptide 8h;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 10 |
|
PDB 1kwi chain A
Region: 4 - 17
Aligned: 14
Modelled: 14
Confidence: 10.9%
Identity: 21%
Fold: Cystatin-like
Superfamily: Cystatin/monellin
Family: Cathelicidin motif
Phyre2
| 11 |
|
PDB 2jo1 chain A
Region: 24 - 42
Aligned: 19
Modelled: 19
Confidence: 7.6%
Identity: 21%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 12 |
|
PDB 2bh1 chain A domain 2
Region: 6 - 22
Aligned: 17
Modelled: 17
Confidence: 7.2%
Identity: 12%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Cyto-EpsL domain
Phyre2
| 13 |
|
PDB 3ez3 chain A
Region: 2 - 26
Aligned: 25
Modelled: 25
Confidence: 6.7%
Identity: 8%
PDB header:lyase
Chain: A: PDB Molecule:farnesyl pyrophosphate synthase, putative;
PDBTitle: crystal structure of plasmodium vivax geranylgeranylpyrophosphate2 synthase pvx_092040 with zoledronate and ipp bound
Phyre2
| 14 |
|
PDB 3db3 chain A
Region: 1 - 15
Aligned: 15
Modelled: 14
Confidence: 6.2%
Identity: 40%
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase uhrf1;
PDBTitle: crystal structure of the tandem tudor domains of the e3 ubiquitin-2 protein ligase uhrf1 in complex with trimethylated histone h3-k93 peptide
Phyre2
| 15 |
|
PDB 3lsn chain A
Region: 2 - 26
Aligned: 25
Modelled: 24
Confidence: 5.5%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:geranyltranstransferase;
PDBTitle: crystal structure of putative geranyltranstransferase from pseudomonas2 fluorescens pf-5 complexed with magnesium
Phyre2
| 16 |
|
PDB 2jp3 chain A
Region: 24 - 42
Aligned: 19
Modelled: 19
Confidence: 5.4%
Identity: 26%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2