| 1 |
|
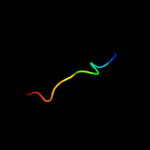
PDB 3a69 chain A
Region: 97 - 136
Aligned: 40
Modelled: 40
Confidence: 98.7%
Identity: 13%
PDB header:motor protein
Chain: A: PDB Molecule:flagellar hook protein flge;
PDBTitle: atomic model of the bacterial flagellar hook based on2 docking an x-ray derived structure and terminal two alpha-3 helices into an 7.1 angstrom resolution cryoem map
Phyre2
| 2 |
|
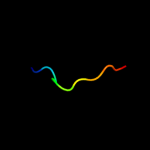
PDB 1ucu chain A
Region: 99 - 136
Aligned: 38
Modelled: 38
Confidence: 90.4%
Identity: 21%
Fold: Phase 1 flagellin
Superfamily: Phase 1 flagellin
Family: Phase 1 flagellin
Phyre2
| 3 |
|
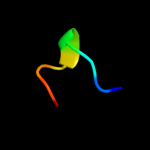
PDB 1ory chain B
Region: 99 - 136
Aligned: 38
Modelled: 38
Confidence: 88.2%
Identity: 26%
PDB header:chaperone
Chain: B: PDB Molecule:flagellin;
PDBTitle: flagellar export chaperone in complex with its cognate binding partner
Phyre2
| 4 |
|
PDB 3k8v chain B
Region: 99 - 119
Aligned: 21
Modelled: 21
Confidence: 44.7%
Identity: 14%
PDB header:structural protein
Chain: B: PDB Molecule:flagellin homolog;
PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
Phyre2
| 5 |
|
PDB 1r6f chain A
Region: 102 - 136
Aligned: 35
Modelled: 35
Confidence: 39.3%
Identity: 23%
Fold: Virulence-associated V antigen
Superfamily: Virulence-associated V antigen
Family: Virulence-associated V antigen
Phyre2
| 6 |
|
PDB 1lvo chain A
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 21.7%
Identity: 36%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 7 |
|
PDB 2q6f chain B
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 19.5%
Identity: 55%
PDB header:hydrolase
Chain: B: PDB Molecule:infectious bronchitis virus (ibv) main protease;
PDBTitle: crystal structure of infectious bronchitis virus (ibv) main2 protease in complex with a michael acceptor inhibitor n3
Phyre2
| 8 |
|
PDB 3d23 chain A
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 16.0%
Identity: 36%
PDB header:hydrolase
Chain: A: PDB Molecule:3c-like proteinase;
PDBTitle: main protease of hcov-hku1
Phyre2
| 9 |
|
PDB 1p9s chain A
Region: 29 - 39
Aligned: 11
Modelled: 10
Confidence: 15.0%
Identity: 27%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 10 |
|
PDB 2duc chain A domain 1
Region: 29 - 39
Aligned: 11
Modelled: 11
Confidence: 14.4%
Identity: 36%
Fold: Trypsin-like serine proteases
Superfamily: Trypsin-like serine proteases
Family: Viral cysteine protease of trypsin fold
Phyre2
| 11 |
|
PDB 1nrj chain A
Region: 30 - 38
Aligned: 9
Modelled: 9
Confidence: 8.0%
Identity: 56%
Fold: Profilin-like
Superfamily: SNARE-like
Family: SRP alpha N-terminal domain-like
Phyre2