| 1 |
|
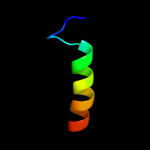
PDB 2qsb chain A domain 1
Region: 25 - 59
Aligned: 35
Modelled: 35
Confidence: 57.9%
Identity: 26%
Fold: Bromodomain-like
Superfamily: Ta0600-like
Family: Ta0600-like
Phyre2
| 2 |
|
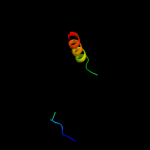
PDB 2qzg chain A domain 1
Region: 25 - 59
Aligned: 35
Modelled: 35
Confidence: 52.3%
Identity: 26%
Fold: Bromodomain-like
Superfamily: Ta0600-like
Family: Ta0600-like
Phyre2
| 3 |
|
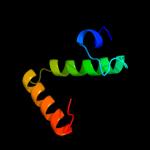
PDB 3lvy chain B
Region: 5 - 64
Aligned: 60
Modelled: 60
Confidence: 27.7%
Identity: 18%
PDB header:lyase
Chain: B: PDB Molecule:carboxymuconolactone decarboxylase family;
PDBTitle: crystal structure of carboxymuconolactone decarboxylase2 family protein smu.961 from streptococcus mutans
Phyre2
| 4 |
|
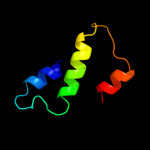
PDB 1x3l chain A
Region: 15 - 49
Aligned: 35
Modelled: 35
Confidence: 22.9%
Identity: 26%
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein ph0495;
PDBTitle: crystal structure of the ph0495 protein from pyrococccus horikoshii2 ot3
Phyre2
| 5 |
|
PDB 3ggz chain C
Region: 1 - 34
Aligned: 34
Modelled: 34
Confidence: 18.1%
Identity: 29%
PDB header:protein transport, endocytosis
Chain: C: PDB Molecule:increased sodium tolerance protein 1;
PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
Phyre2
| 6 |
|
PDB 2rg8 chain A
Region: 23 - 61
Aligned: 39
Modelled: 39
Confidence: 17.2%
Identity: 31%
PDB header:apoptosis, translation
Chain: A: PDB Molecule:programmed cell death protein 4;
PDBTitle: crystal structure of programmed for cell death 4 middle ma32 domain
Phyre2
| 7 |
|
PDB 2kxh chain B
Region: 18 - 39
Aligned: 22
Modelled: 22
Confidence: 12.7%
Identity: 41%
PDB header:protein binding
Chain: B: PDB Molecule:peptide of far upstream element-binding protein 1;
PDBTitle: solution structure of the first two rrm domains of fir in the complex2 with fbp nbox peptide
Phyre2
| 8 |
|
PDB 2p02 chain A domain 2
Region: 14 - 38
Aligned: 25
Modelled: 25
Confidence: 12.6%
Identity: 32%
Fold: S-adenosylmethionine synthetase
Superfamily: S-adenosylmethionine synthetase
Family: S-adenosylmethionine synthetase
Phyre2
| 9 |
|
PDB 1qm4 chain A domain 2
Region: 14 - 38
Aligned: 25
Modelled: 25
Confidence: 11.5%
Identity: 32%
Fold: S-adenosylmethionine synthetase
Superfamily: S-adenosylmethionine synthetase
Family: S-adenosylmethionine synthetase
Phyre2
| 10 |
|
PDB 2ouw chain A domain 1
Region: 8 - 63
Aligned: 56
Modelled: 56
Confidence: 10.1%
Identity: 20%
Fold: AhpD-like
Superfamily: AhpD-like
Family: TTHA0727-like
Phyre2
| 11 |
|
PDB 3fp5 chain A
Region: 1 - 17
Aligned: 17
Modelled: 17
Confidence: 10.1%
Identity: 18%
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl-coa binding protein;
PDBTitle: crystal structure of acbp from moniliophthora perniciosa
Phyre2
| 12 |
|
PDB 1mxa chain A domain 2
Region: 14 - 38
Aligned: 25
Modelled: 25
Confidence: 9.6%
Identity: 28%
Fold: S-adenosylmethionine synthetase
Superfamily: S-adenosylmethionine synthetase
Family: S-adenosylmethionine synthetase
Phyre2
| 13 |
|
PDB 3c1l chain B
Region: 20 - 64
Aligned: 45
Modelled: 45
Confidence: 9.2%
Identity: 20%
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative antioxidant defense protein mlr4105;
PDBTitle: crystal structure of an antioxidant defense protein (mlr4105) from2 mesorhizobium loti maff303099 at 2.00 a resolution
Phyre2
| 14 |
|
PDB 2j9w chain B
Region: 5 - 53
Aligned: 49
Modelled: 49
Confidence: 8.0%
Identity: 16%
PDB header:protein transport
Chain: B: PDB Molecule:vps28-prov protein;
PDBTitle: structural insight into the escrt-i-ii link and its role in2 mvb trafficking
Phyre2
| 15 |
|
PDB 1s5a chain A
Region: 1 - 20
Aligned: 20
Modelled: 12
Confidence: 7.3%
Identity: 40%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: PhzA/PhzB-like
Phyre2
| 16 |
|
PDB 3ix6 chain B
Region: 1 - 14
Aligned: 14
Modelled: 14
Confidence: 6.8%
Identity: 21%
PDB header:transferase
Chain: B: PDB Molecule:thymidylate synthase;
PDBTitle: crystal structure of thymidylate synthase thya from brucella2 melitensis
Phyre2
| 17 |
|
PDB 2pfx chain A domain 1
Region: 20 - 64
Aligned: 45
Modelled: 45
Confidence: 5.9%
Identity: 18%
Fold: AhpD-like
Superfamily: AhpD-like
Family: Atu0492-like
Phyre2
| 18 |
|
PDB 2cqu chain A
Region: 1 - 21
Aligned: 21
Modelled: 21
Confidence: 5.7%
Identity: 14%
PDB header:isomerase
Chain: A: PDB Molecule:peroxisomal d3,d2-enoyl-coa isomerase;
PDBTitle: solution structure of rsgi ruh-045, a human acyl-coa2 binding protein
Phyre2
| 19 |
|
PDB 2gy9 chain T domain 1
Region: 7 - 46
Aligned: 40
Modelled: 40
Confidence: 5.5%
Identity: 25%
Fold: Spectrin repeat-like
Superfamily: Ribosomal protein S20
Family: Ribosomal protein S20
Phyre2
| 20 |
|
PDB 2w3c chain A
Region: 30 - 59
Aligned: 30
Modelled: 30
Confidence: 5.4%
Identity: 7%
PDB header:transport protein
Chain: A: PDB Molecule:general vesicular transport factor p115;
PDBTitle: globular head region of the human general vesicular2 transport factor p115
Phyre2
| 21 |
|