| 1 |
|
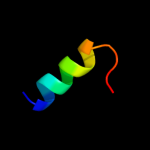
PDB 1cii chain A
Region: 36 - 75
Aligned: 40
Modelled: 40
Confidence: 64.1%
Identity: 20%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 2 |
|
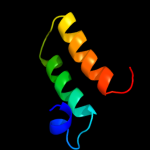
PDB 3aqp chain B
Region: 12 - 77
Aligned: 66
Modelled: 66
Confidence: 22.1%
Identity: 27%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 3 |
|
PDB 1qg9 chain A
Region: 61 - 78
Aligned: 18
Modelled: 16
Confidence: 18.9%
Identity: 28%
PDB header:transmembrane channel
Chain: A: PDB Molecule:protein (sodium channel protein, brain ii alpha
PDBTitle: second repeat (is2mic) from voltage-gated sodium channel
Phyre2
| 4 |
|
PDB 1a8r chain A
Region: 50 - 77
Aligned: 28
Modelled: 28
Confidence: 18.0%
Identity: 14%
Fold: T-fold
Superfamily: Tetrahydrobiopterin biosynthesis enzymes-like
Family: GTP cyclohydrolase I
Phyre2
| 5 |
|
PDB 1wpl chain A
Region: 50 - 77
Aligned: 28
Modelled: 28
Confidence: 16.3%
Identity: 39%
Fold: T-fold
Superfamily: Tetrahydrobiopterin biosynthesis enzymes-like
Family: GTP cyclohydrolase I
Phyre2
| 6 |
|
PDB 1is7 chain F
Region: 50 - 77
Aligned: 28
Modelled: 28
Confidence: 14.7%
Identity: 39%
PDB header:hydrolase/protein binding
Chain: F: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: crystal structure of rat gtpchi/gfrp stimulatory complex
Phyre2
| 7 |
|
PDB 3jzd chain A
Region: 22 - 78
Aligned: 56
Modelled: 57
Confidence: 13.1%
Identity: 14%
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-containing alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehedrogenase (yp_298327.1) from2 ralstonia eutropha jmp134 at 2.10 a resolution
Phyre2
| 8 |
|
PDB 1jo5 chain A
Region: 60 - 76
Aligned: 17
Modelled: 17
Confidence: 13.1%
Identity: 18%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 9 |
|
PDB 1wur chain A domain 1
Region: 50 - 77
Aligned: 28
Modelled: 28
Confidence: 12.8%
Identity: 39%
Fold: T-fold
Superfamily: Tetrahydrobiopterin biosynthesis enzymes-like
Family: GTP cyclohydrolase I
Phyre2
| 10 |
|
PDB 1wm9 chain D
Region: 50 - 77
Aligned: 28
Modelled: 28
Confidence: 12.8%
Identity: 39%
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: structure of gtp cyclohydrolase i from thermus thermophilus hb8
Phyre2
| 11 |
|
PDB 1ztd chain A domain 1
Region: 47 - 59
Aligned: 13
Modelled: 13
Confidence: 12.1%
Identity: 62%
Fold: RNase III domain-like
Superfamily: RNase III domain-like
Family: PF0609-like
Phyre2
| 12 |
|
PDB 2xzm chain O
Region: 9 - 30
Aligned: 22
Modelled: 22
Confidence: 9.7%
Identity: 27%
PDB header:ribosome
Chain: O: PDB Molecule:rps13e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 13 |
|
PDB 1rh1 chain A domain 2
Region: 32 - 61
Aligned: 30
Modelled: 30
Confidence: 9.6%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 14 |
|
PDB 2ari chain A
Region: 40 - 51
Aligned: 12
Modelled: 12
Confidence: 9.4%
Identity: 58%
PDB header:viral protein
Chain: A: PDB Molecule:envelope polyprotein gp160;
PDBTitle: solution structure of micelle-bound fusion domain of hiv-12 gp41
Phyre2
| 15 |
|
PDB 1h2d chain A
Region: 20 - 56
Aligned: 37
Modelled: 37
Confidence: 9.2%
Identity: 30%
PDB header:virus/viral protein
Chain: A: PDB Molecule:matrix protein vp40;
PDBTitle: ebola virus matrix protein vp40 n-terminal domain in2 complex with rna (low-resolution vp40[31-212] variant).
Phyre2
| 16 |
|
PDB 1h2c chain A
Region: 32 - 56
Aligned: 25
Modelled: 25
Confidence: 9.0%
Identity: 28%
Fold: EV matrix protein
Superfamily: EV matrix protein
Family: EV matrix protein
Phyre2
| 17 |
|
PDB 3izb chain O
Region: 9 - 30
Aligned: 22
Modelled: 22
Confidence: 8.6%
Identity: 14%
PDB header:ribosome
Chain: O: PDB Molecule:40s ribosomal protein rps13 (s15p);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 18 |
|
PDB 1cii chain A domain 1
Region: 32 - 75
Aligned: 44
Modelled: 44
Confidence: 7.6%
Identity: 18%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 19 |
|
PDB 1wrg chain A
Region: 60 - 76
Aligned: 17
Modelled: 17
Confidence: 7.1%
Identity: 29%
PDB header:membrane protein
Chain: A: PDB Molecule:light-harvesting protein b-880, beta chain;
PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
Phyre2
| 20 |
|
PDB 2i88 chain A
Region: 32 - 77
Aligned: 46
Modelled: 46
Confidence: 6.9%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:colicin-e1;
PDBTitle: crystal structure of the channel-forming domain of colicin2 e1
Phyre2
| 21 |
|